| Title: | Utilities for Scoring and Assessing Predictions |
| Version: | 2.1.2 |
| Language: | en-GB |
| Description: | Facilitate the evaluation of forecasts in a convenient framework based on data.table. It allows user to to check their forecasts and diagnose issues, to visualise forecasts and missing data, to transform data before scoring, to handle missing forecasts, to aggregate scores, and to visualise the results of the evaluation. The package mostly focuses on the evaluation of probabilistic forecasts and allows evaluating several different forecast types and input formats. Find more information about the package in the Vignettes as well as in the accompanying paper, <doi:10.48550/arXiv.2205.07090>. |
| License: | MIT + file LICENSE |
| Encoding: | UTF-8 |
| LazyData: | true |
| Imports: | checkmate, cli, data.table (≥ 1.16.0), ggplot2 (≥ 3.4.0), methods, Metrics, purrr, scoringRules (≥ 1.1.3), stats |
| Suggests: | ggdist, kableExtra, knitr, magrittr, rmarkdown, testthat (≥ 3.1.9), vdiffr |
| Config/Needs/website: | r-lib/pkgdown, amirmasoudabdol/preferably |
| Config/testthat/edition: | 3 |
| RoxygenNote: | 7.3.2 |
| URL: | https://doi.org/10.48550/arXiv.2205.07090, https://epiforecasts.io/scoringutils/, https://github.com/epiforecasts/scoringutils |
| BugReports: | https://github.com/epiforecasts/scoringutils/issues |
| VignetteBuilder: | knitr |
| Depends: | R (≥ 4.1) |
| NeedsCompilation: | no |
| Packaged: | 2025-08-25 07:24:53 UTC; nikos |
| Author: | Nikos Bosse |
| Maintainer: | Nikos Bosse <nikosbosse@gmail.com> |
| Repository: | CRAN |
| Date/Publication: | 2025-08-25 08:10:02 UTC |
scoringutils: Utilities for Scoring and Assessing Predictions
Description
Facilitate the evaluation of forecasts in a convenient framework based on data.table. It allows user to to check their forecasts and diagnose issues, to visualise forecasts and missing data, to transform data before scoring, to handle missing forecasts, to aggregate scores, and to visualise the results of the evaluation. The package mostly focuses on the evaluation of probabilistic forecasts and allows evaluating several different forecast types and input formats. Find more information about the package in the Vignettes as well as in the accompanying paper, doi:10.48550/arXiv.2205.07090.
Author(s)
Maintainer: Nikos Bosse nikosbosse@gmail.com (ORCID)
Authors:
Sam Abbott contact@samabbott.co.uk (ORCID)
Hugo Gruson hugo.gruson+R@normalesup.org (ORCID)
Sebastian Funk sebastian.funk@lshtm.ac.uk
Other contributors:
Johannes Bracher johannes.bracher@kit.edu (ORCID) [contributor]
Toshiaki Asakura toshiaki.asa9ra@gmail.com (ORCID) [contributor]
James Mba Azam james.azam@lshtm.ac.uk (ORCID) [contributor]
Michael Chirico michaelchirico4@gmail.com (ORCID) [contributor]
See Also
Useful links:
Report bugs at https://github.com/epiforecasts/scoringutils/issues
Add relative skill scores based on pairwise comparisons
Description
Adds a columns with relative skills computed by running
pairwise comparisons on the scores.
For more information on
the computation of relative skill, see get_pairwise_comparisons().
Relative skill will be calculated for the aggregation level specified in
by.
Usage
add_relative_skill(
scores,
compare = "model",
by = NULL,
metric = intersect(c("wis", "crps", "brier_score"), names(scores)),
baseline = NULL,
...
)
Arguments
scores |
An object of class |
compare |
Character vector with a single colum name that defines the elements for the pairwise comparison. For example, if this is set to "model" (the default), then elements of the "model" column will be compared. |
by |
Character vector with column names that define further grouping
levels for the pairwise comparisons. By default this is |
metric |
A string with the name of the metric for which a relative skill shall be computed. By default this is either "crps", "wis" or "brier_score" if any of these are available. |
baseline |
A string with the name of a model. If a baseline is
given, then a scaled relative skill with respect to the baseline will be
returned. By default ( |
... |
Additional arguments for the comparison between two models. See
|
Absolute error of the median (quantile-based version)
Description
Compute the absolute error of the median calculated as
|\text{observed} - \text{median prediction}|
The median prediction is the predicted value for which quantile_level == 0.5.
The function requires 0.5 to be among the quantile levels in quantile_level.
Usage
ae_median_quantile(observed, predicted, quantile_level)
Arguments
observed |
Numeric vector of size n with the observed values. |
predicted |
Numeric nxN matrix of predictive
quantiles, n (number of rows) being the number of forecasts (corresponding
to the number of observed values) and N
(number of columns) the number of quantiles per forecast.
If |
quantile_level |
Vector of of size N with the quantile levels for which predictions were made. |
Value
Numeric vector of length N with the absolute error of the median.
Input format
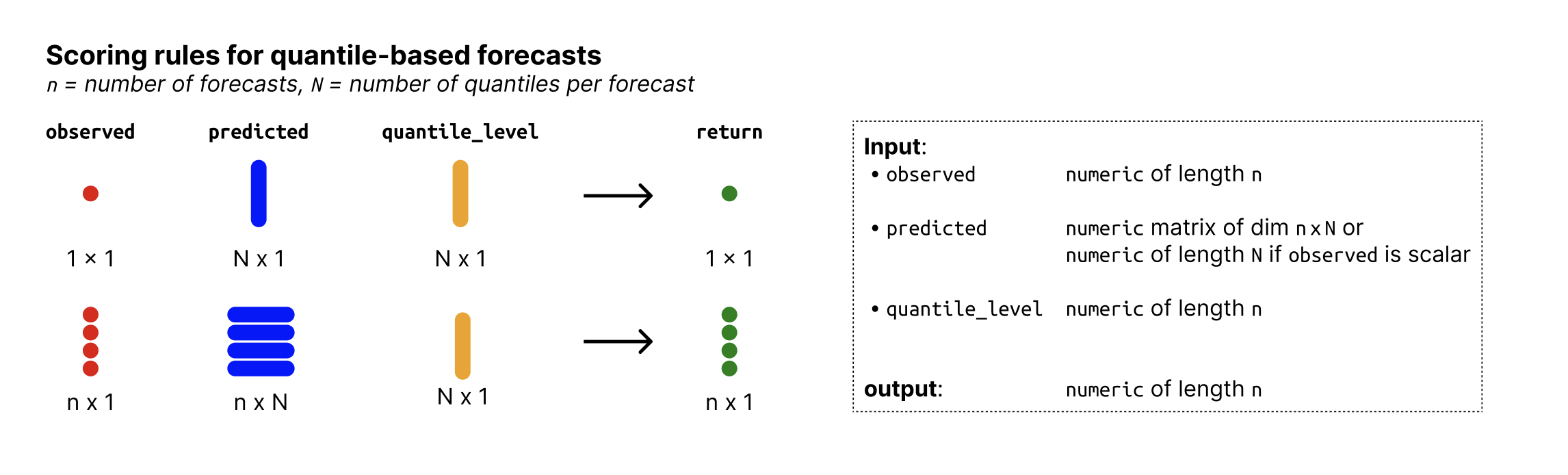
Overview of required input format for quantile-based forecasts
See Also
Examples
observed <- rnorm(30, mean = 1:30)
predicted_values <- replicate(3, rnorm(30, mean = 1:30))
ae_median_quantile(
observed, predicted_values, quantile_level = c(0.2, 0.5, 0.8)
)
Absolute error of the median (sample-based version)
Description
Absolute error of the median calculated as
|\text{observed} - \text{median prediction}|
where the median prediction is calculated as the median of the predictive samples.
Usage
ae_median_sample(observed, predicted)
Arguments
observed |
A vector with observed values of size n |
predicted |
nxN matrix of predictive samples, n (number of rows) being
the number of data points and N (number of columns) the number of Monte
Carlo samples. Alternatively, |
Value
Numeric vector of length n with the absolute errors of the median.
Input format
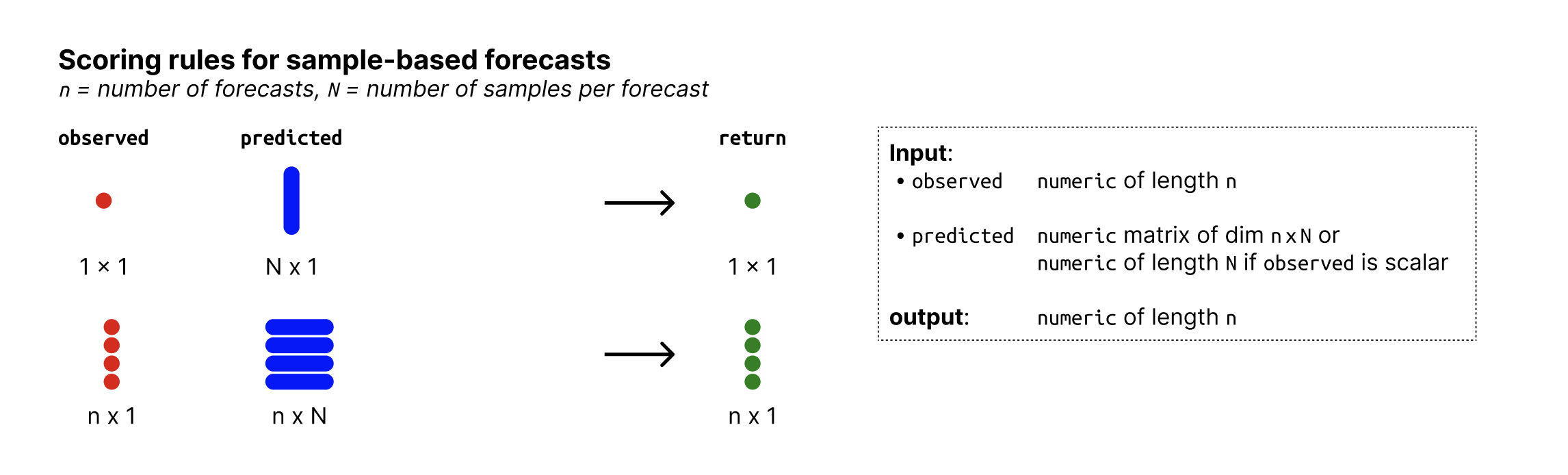
Overview of required input format for sample-based forecasts
See Also
Examples
observed <- rnorm(30, mean = 1:30)
predicted_values <- matrix(rnorm(30, mean = 1:30))
ae_median_sample(observed, predicted_values)
Apply a list of functions to a data table of forecasts
Description
This helper function applies scoring rules (stored as a list of
functions) to a data table of forecasts. apply_metrics is used within
score() to apply all scoring rules to the data.
Scoring rules are wrapped in run_safely() to catch errors and to make
sure that only arguments are passed to the scoring rule that are actually
accepted by it.
Usage
apply_metrics(forecast, metrics, ...)
Arguments
forecast |
A forecast object (a validated data.table with predicted and observed values). |
metrics |
A named list of scoring functions. Names will be used as
column names in the output. See |
... |
Additional arguments to be passed to the scoring rules. Note that
this is currently not used, as all calls to |
Value
A data table with the forecasts and the calculated metrics.
Create a forecast object for binary forecasts
Description
Process and validate a data.frame (or similar) or similar with forecasts
and observations. If the input passes all input checks, those functions will
be converted to a forecast object. A forecast object is a data.table with
a class forecast and an additional class that depends on the forecast type.
The arguments observed, predicted, etc. make it possible to rename
existing columns of the input data to match the required columns for a
forecast object. Using the argument forecast_unit, you can specify
the columns that uniquely identify a single forecast (and thereby removing
other, unneeded columns. See section "Forecast Unit" below for details).
Usage
as_forecast_binary(data, ...)
## Default S3 method:
as_forecast_binary(
data,
forecast_unit = NULL,
observed = NULL,
predicted = NULL,
...
)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
... |
Unused |
forecast_unit |
(optional) Name of the columns in |
observed |
(optional) Name of the column in |
predicted |
(optional) Name of the column in |
Value
A forecast object of class forecast_binary
Required input
The input needs to be a data.frame or similar with the following columns:
-
observed:factorwith exactly two levels representing the observed values. The highest factor level is assumed to be the reference level. This means that corresponding value inpredictedrepresent the probability that the observed value is equal to the highest factor level. -
predicted:numericwith predicted probabilities, representing the probability that the corresponding value inobservedis equal to the highest available factor level.
For convenience, we recommend an additional column model holding the name
of the forecaster or model that produced a prediction, but this is not
strictly necessary.
See the example_binary data set for an example.
Forecast unit
In order to score forecasts, scoringutils needs to know which of the rows
of the data belong together and jointly form a single forecasts. This is
easy e.g. for point forecast, where there is one row per forecast. For
quantile or sample-based forecasts, however, there are multiple rows that
belong to a single forecast.
The forecast unit or unit of a single forecast is then described by the
combination of columns that uniquely identify a single forecast.
For example, we could have forecasts made by different models in various
locations at different time points, each for several weeks into the future.
The forecast unit could then be described as
forecast_unit = c("model", "location", "forecast_date", "forecast_horizon").
scoringutils automatically tries to determine the unit of a single
forecast. It uses all existing columns for this, which means that no columns
must be present that are unrelated to the forecast unit. As a very simplistic
example, if you had an additional row, "even", that is one if the row number
is even and zero otherwise, then this would mess up scoring as scoringutils
then thinks that this column was relevant in defining the forecast unit.
In order to avoid issues, we recommend setting the forecast unit explicitly,
using the forecast_unit argument. This will simply drop unneeded columns,
while making sure that all necessary, 'protected columns' like "predicted"
or "observed" are retained.
See Also
Other functions to create forecast objects:
as_forecast_nominal(),
as_forecast_ordinal(),
as_forecast_point(),
as_forecast_quantile(),
as_forecast_sample()
Examples
as_forecast_binary(
example_binary,
predicted = "predicted",
forecast_unit = c("model", "target_type", "target_end_date",
"horizon", "location")
)
General information on creating a forecast object
Description
Process and validate a data.frame (or similar) or similar with forecasts
and observations. If the input passes all input checks, those functions will
be converted to a forecast object. A forecast object is a data.table with
a class forecast and an additional class that depends on the forecast type.
The arguments observed, predicted, etc. make it possible to rename
existing columns of the input data to match the required columns for a
forecast object. Using the argument forecast_unit, you can specify
the columns that uniquely identify a single forecast (and thereby removing
other, unneeded columns. See section "Forecast Unit" below for details).
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
forecast_unit |
(optional) Name of the columns in |
observed |
(optional) Name of the column in |
predicted |
(optional) Name of the column in |
Forecast unit
In order to score forecasts, scoringutils needs to know which of the rows
of the data belong together and jointly form a single forecasts. This is
easy e.g. for point forecast, where there is one row per forecast. For
quantile or sample-based forecasts, however, there are multiple rows that
belong to a single forecast.
The forecast unit or unit of a single forecast is then described by the
combination of columns that uniquely identify a single forecast.
For example, we could have forecasts made by different models in various
locations at different time points, each for several weeks into the future.
The forecast unit could then be described as
forecast_unit = c("model", "location", "forecast_date", "forecast_horizon").
scoringutils automatically tries to determine the unit of a single
forecast. It uses all existing columns for this, which means that no columns
must be present that are unrelated to the forecast unit. As a very simplistic
example, if you had an additional row, "even", that is one if the row number
is even and zero otherwise, then this would mess up scoring as scoringutils
then thinks that this column was relevant in defining the forecast unit.
In order to avoid issues, we recommend setting the forecast unit explicitly,
using the forecast_unit argument. This will simply drop unneeded columns,
while making sure that all necessary, 'protected columns' like "predicted"
or "observed" are retained.
Common functionality for as_forecast_<type> functions
Description
Common functionality for as_forecast_<type> functions
Usage
as_forecast_generic(data, forecast_unit = NULL, ...)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
forecast_unit |
(optional) Name of the columns in |
... |
Named arguments that are used to rename columns. The names of the arguments are the names of the columns that should be renamed. The values are the new names. |
Details
This function splits out part of the functionality of
as_forecast_<type> that is the same for all as_forecast_<type> functions.
It renames the required columns, where appropriate, and sets the forecast
unit.
Create a forecast object for nominal forecasts
Description
Process and validate a data.frame (or similar) or similar with forecasts
and observations. If the input passes all input checks, those functions will
be converted to a forecast object. A forecast object is a data.table with
a class forecast and an additional class that depends on the forecast type.
The arguments observed, predicted, etc. make it possible to rename
existing columns of the input data to match the required columns for a
forecast object. Using the argument forecast_unit, you can specify
the columns that uniquely identify a single forecast (and thereby removing
other, unneeded columns. See section "Forecast Unit" below for details).
Usage
as_forecast_nominal(data, ...)
## Default S3 method:
as_forecast_nominal(
data,
forecast_unit = NULL,
observed = NULL,
predicted = NULL,
predicted_label = NULL,
...
)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
... |
Unused |
forecast_unit |
(optional) Name of the columns in |
observed |
(optional) Name of the column in |
predicted |
(optional) Name of the column in |
predicted_label |
(optional) Name of the column in |
Details
Nominal forecasts are a form of categorical forecasts and represent a generalisation of binary forecasts to multiple outcomes. The possible outcomes that the observed values can assume are not ordered.
Value
A forecast object of class forecast_nominal
Required input
The input needs to be a data.frame or similar for the default method with the following columns:
-
observed: Column with observed values of typefactorwith N levels, where N is the number of possible outcomes. The levels of the factor represent the possible outcomes that the observed values can assume. -
predicted:numericcolumn with predicted probabilities. The values represent the probability that the observed value is equal to the factor level denoted inpredicted_label. Note that forecasts must be complete, i.e. there must be a probability assigned to every possible outcome and those probabilities must sum to one. -
predicted_label:factorwith N levels, denoting the outcome that the probabilities inpredictedcorrespond to.
For convenience, we recommend an additional column model holding the name
of the forecaster or model that produced a prediction, but this is not
strictly necessary.
See the example_nominal data set for an example.
Forecast unit
In order to score forecasts, scoringutils needs to know which of the rows
of the data belong together and jointly form a single forecasts. This is
easy e.g. for point forecast, where there is one row per forecast. For
quantile or sample-based forecasts, however, there are multiple rows that
belong to a single forecast.
The forecast unit or unit of a single forecast is then described by the
combination of columns that uniquely identify a single forecast.
For example, we could have forecasts made by different models in various
locations at different time points, each for several weeks into the future.
The forecast unit could then be described as
forecast_unit = c("model", "location", "forecast_date", "forecast_horizon").
scoringutils automatically tries to determine the unit of a single
forecast. It uses all existing columns for this, which means that no columns
must be present that are unrelated to the forecast unit. As a very simplistic
example, if you had an additional row, "even", that is one if the row number
is even and zero otherwise, then this would mess up scoring as scoringutils
then thinks that this column was relevant in defining the forecast unit.
In order to avoid issues, we recommend setting the forecast unit explicitly,
using the forecast_unit argument. This will simply drop unneeded columns,
while making sure that all necessary, 'protected columns' like "predicted"
or "observed" are retained.
See Also
Other functions to create forecast objects:
as_forecast_binary(),
as_forecast_ordinal(),
as_forecast_point(),
as_forecast_quantile(),
as_forecast_sample()
Examples
as_forecast_nominal(
na.omit(example_nominal),
predicted = "predicted",
forecast_unit = c("model", "target_type", "target_end_date",
"horizon", "location")
)
Create a forecast object for ordinal forecasts
Description
Process and validate a data.frame (or similar) or similar with forecasts
and observations. If the input passes all input checks, those functions will
be converted to a forecast object. A forecast object is a data.table with
a class forecast and an additional class that depends on the forecast type.
The arguments observed, predicted, etc. make it possible to rename
existing columns of the input data to match the required columns for a
forecast object. Using the argument forecast_unit, you can specify
the columns that uniquely identify a single forecast (and thereby removing
other, unneeded columns. See section "Forecast Unit" below for details).
Usage
as_forecast_ordinal(data, ...)
## Default S3 method:
as_forecast_ordinal(
data,
forecast_unit = NULL,
observed = NULL,
predicted = NULL,
predicted_label = NULL,
...
)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
... |
Unused |
forecast_unit |
(optional) Name of the columns in |
observed |
(optional) Name of the column in |
predicted |
(optional) Name of the column in |
predicted_label |
(optional) Name of the column in |
Details
Ordinal forecasts are a form of categorical forecasts and represent a generalisation of binary forecasts to multiple outcomes. The possible outcomes that the observed values can assume are ordered.
Value
A forecast object of class forecast_ordinal
Required input
The input needs to be a data.frame or similar for the default method with the following columns:
-
observed: Column with observed values of typefactorwith N ordered levels, where N is the number of possible outcomes. The levels of the factor represent the possible outcomes that the observed values can assume. -
predicted:numericcolumn with predicted probabilities. The values represent the probability that the observed value is equal to the factor level denoted inpredicted_label. Note that forecasts must be complete, i.e. there must be a probability assigned to every possible outcome and those probabilities must sum to one. -
predicted_label:factorwith N levels, denoting the outcome that the probabilities inpredictedcorrespond to.
For convenience, we recommend an additional column model holding the name
of the forecaster or model that produced a prediction, but this is not
strictly necessary.
See the example_ordinal data set for an example.
Forecast unit
In order to score forecasts, scoringutils needs to know which of the rows
of the data belong together and jointly form a single forecasts. This is
easy e.g. for point forecast, where there is one row per forecast. For
quantile or sample-based forecasts, however, there are multiple rows that
belong to a single forecast.
The forecast unit or unit of a single forecast is then described by the
combination of columns that uniquely identify a single forecast.
For example, we could have forecasts made by different models in various
locations at different time points, each for several weeks into the future.
The forecast unit could then be described as
forecast_unit = c("model", "location", "forecast_date", "forecast_horizon").
scoringutils automatically tries to determine the unit of a single
forecast. It uses all existing columns for this, which means that no columns
must be present that are unrelated to the forecast unit. As a very simplistic
example, if you had an additional row, "even", that is one if the row number
is even and zero otherwise, then this would mess up scoring as scoringutils
then thinks that this column was relevant in defining the forecast unit.
In order to avoid issues, we recommend setting the forecast unit explicitly,
using the forecast_unit argument. This will simply drop unneeded columns,
while making sure that all necessary, 'protected columns' like "predicted"
or "observed" are retained.
See Also
Other functions to create forecast objects:
as_forecast_binary(),
as_forecast_nominal(),
as_forecast_point(),
as_forecast_quantile(),
as_forecast_sample()
Examples
as_forecast_ordinal(
na.omit(example_ordinal),
predicted = "predicted",
forecast_unit = c("model", "target_type", "target_end_date",
"horizon", "location")
)
Create a forecast object for point forecasts
Description
When converting a forecast_quantile object into a forecast_point object,
the 0.5 quantile is extracted and returned as the point forecast.
Usage
as_forecast_point(data, ...)
## Default S3 method:
as_forecast_point(
data,
forecast_unit = NULL,
observed = NULL,
predicted = NULL,
...
)
## S3 method for class 'forecast_quantile'
as_forecast_point(data, ...)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
... |
Unused |
forecast_unit |
(optional) Name of the columns in |
observed |
(optional) Name of the column in |
predicted |
(optional) Name of the column in |
Value
A forecast object of class forecast_point
Required input
The input needs to be a data.frame or similar for the default method with the following columns:
-
observed: Column of typenumericwith observed values. -
predicted: Column of typenumericwith predicted values.
For convenience, we recommend an additional column model holding the name
of the forecaster or model that produced a prediction, but this is not
strictly necessary.
See the example_point data set for an example.
See Also
Other functions to create forecast objects:
as_forecast_binary(),
as_forecast_nominal(),
as_forecast_ordinal(),
as_forecast_quantile(),
as_forecast_sample()
Create a forecast object for quantile-based forecasts
Description
Process and validate a data.frame (or similar) or similar with forecasts
and observations. If the input passes all input checks, those functions will
be converted to a forecast object. A forecast object is a data.table with
a class forecast and an additional class that depends on the forecast type.
The arguments observed, predicted, etc. make it possible to rename
existing columns of the input data to match the required columns for a
forecast object. Using the argument forecast_unit, you can specify
the columns that uniquely identify a single forecast (and thereby removing
other, unneeded columns. See section "Forecast Unit" below for details).
Usage
as_forecast_quantile(data, ...)
## Default S3 method:
as_forecast_quantile(
data,
forecast_unit = NULL,
observed = NULL,
predicted = NULL,
quantile_level = NULL,
...
)
## S3 method for class 'forecast_sample'
as_forecast_quantile(
data,
probs = c(0.05, 0.25, 0.5, 0.75, 0.95),
type = 7,
...
)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
... |
Unused |
forecast_unit |
(optional) Name of the columns in |
observed |
(optional) Name of the column in |
predicted |
(optional) Name of the column in |
quantile_level |
(optional) Name of the column in |
probs |
A numeric vector of quantile levels for which
quantiles will be computed. Corresponds to the |
type |
Type argument passed down to the quantile function. For more
information, see |
Value
A forecast object of class forecast_quantile
Required input
The input needs to be a data.frame or similar for the default method with the following columns:
-
observed: Column of typenumericwith observed values. -
predicted: Column of typenumericwith predicted values. Predicted values represent quantiles of the predictive distribution. -
quantile_level: Column of typenumeric, denoting the quantile level of the corresponding predicted value. Quantile levels must be between 0 and 1.
For convenience, we recommend an additional column model holding the name
of the forecaster or model that produced a prediction, but this is not
strictly necessary.
See the example_quantile data set for an example.
Converting from forecast_sample to forecast_quantile
When creating a forecast_quantile object from a forecast_sample object,
the quantiles are estimated by computing empircal quantiles from the samples
via quantile(). Note that empirical quantiles are a biased estimator for
the true quantiles in particular in the tails of the distribution and
when the number of available samples is low.
Forecast unit
In order to score forecasts, scoringutils needs to know which of the rows
of the data belong together and jointly form a single forecasts. This is
easy e.g. for point forecast, where there is one row per forecast. For
quantile or sample-based forecasts, however, there are multiple rows that
belong to a single forecast.
The forecast unit or unit of a single forecast is then described by the
combination of columns that uniquely identify a single forecast.
For example, we could have forecasts made by different models in various
locations at different time points, each for several weeks into the future.
The forecast unit could then be described as
forecast_unit = c("model", "location", "forecast_date", "forecast_horizon").
scoringutils automatically tries to determine the unit of a single
forecast. It uses all existing columns for this, which means that no columns
must be present that are unrelated to the forecast unit. As a very simplistic
example, if you had an additional row, "even", that is one if the row number
is even and zero otherwise, then this would mess up scoring as scoringutils
then thinks that this column was relevant in defining the forecast unit.
In order to avoid issues, we recommend setting the forecast unit explicitly,
using the forecast_unit argument. This will simply drop unneeded columns,
while making sure that all necessary, 'protected columns' like "predicted"
or "observed" are retained.
See Also
Other functions to create forecast objects:
as_forecast_binary(),
as_forecast_nominal(),
as_forecast_ordinal(),
as_forecast_point(),
as_forecast_sample()
Examples
as_forecast_quantile(
example_quantile,
predicted = "predicted",
forecast_unit = c("model", "target_type", "target_end_date",
"horizon", "location")
)
Create a forecast object for sample-based forecasts
Description
Process and validate a data.frame (or similar) or similar with forecasts
and observations. If the input passes all input checks, those functions will
be converted to a forecast object. A forecast object is a data.table with
a class forecast and an additional class that depends on the forecast type.
The arguments observed, predicted, etc. make it possible to rename
existing columns of the input data to match the required columns for a
forecast object. Using the argument forecast_unit, you can specify
the columns that uniquely identify a single forecast (and thereby removing
other, unneeded columns. See section "Forecast Unit" below for details).
Usage
as_forecast_sample(data, ...)
## Default S3 method:
as_forecast_sample(
data,
forecast_unit = NULL,
observed = NULL,
predicted = NULL,
sample_id = NULL,
...
)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
... |
Unused |
forecast_unit |
(optional) Name of the columns in |
observed |
(optional) Name of the column in |
predicted |
(optional) Name of the column in |
sample_id |
(optional) Name of the column in |
Value
A forecast object of class forecast_sample
Required input
The input needs to be a data.frame or similar for the default method with the following columns:
-
observed: Column of typenumericwith observed values. -
predicted: Column of typenumericwith predicted values. Predicted values represent random samples from the predictive distribution. -
sample_id: Column of any type with unique identifiers (unique within a single forecast) for each sample.
For convenience, we recommend an additional column model holding the name
of the forecaster or model that produced a prediction, but this is not
strictly necessary.
See the example_sample_continuous and example_sample_discrete data set for an example
Forecast unit
In order to score forecasts, scoringutils needs to know which of the rows
of the data belong together and jointly form a single forecasts. This is
easy e.g. for point forecast, where there is one row per forecast. For
quantile or sample-based forecasts, however, there are multiple rows that
belong to a single forecast.
The forecast unit or unit of a single forecast is then described by the
combination of columns that uniquely identify a single forecast.
For example, we could have forecasts made by different models in various
locations at different time points, each for several weeks into the future.
The forecast unit could then be described as
forecast_unit = c("model", "location", "forecast_date", "forecast_horizon").
scoringutils automatically tries to determine the unit of a single
forecast. It uses all existing columns for this, which means that no columns
must be present that are unrelated to the forecast unit. As a very simplistic
example, if you had an additional row, "even", that is one if the row number
is even and zero otherwise, then this would mess up scoring as scoringutils
then thinks that this column was relevant in defining the forecast unit.
In order to avoid issues, we recommend setting the forecast unit explicitly,
using the forecast_unit argument. This will simply drop unneeded columns,
while making sure that all necessary, 'protected columns' like "predicted"
or "observed" are retained.
See Also
Other functions to create forecast objects:
as_forecast_binary(),
as_forecast_nominal(),
as_forecast_ordinal(),
as_forecast_point(),
as_forecast_quantile()
Create an object of class scores from data
Description
This convenience function wraps new_scores() and validates
the scores object.
Usage
as_scores(scores, metrics)
Arguments
scores |
A data.table or similar with scores as produced by |
metrics |
A character vector with the names of the scores (i.e. the names of the scoring rules used for scoring). |
Value
An object of class scores
Assert Inputs Have Matching Dimensions
Description
Function assesses whether input dimensions match. In the following, n is the number of observations / forecasts. Scalar values may be repeated to match the length of the other input. Allowed options are therefore:
-
observedis vector of length 1 or length n -
predictedis:a vector of of length 1 or length n
a matrix with n rows and 1 column
Usage
assert_dims_ok_point(observed, predicted)
Arguments
observed |
Input to be checked. Should be a factor of length n with
exactly two levels, holding the observed values.
The highest factor level is assumed to be the reference level. This means
that |
predicted |
Input to be checked. |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Assert that input is a forecast object and passes validations
Description
Assert that an object is a forecast object (i.e. a data.table with a class
forecast and an additional class forecast_<type> corresponding to the
forecast type).
See the corresponding assert_forecast_<type> functions for more details on
the required input formats.
Usage
## S3 method for class 'forecast_binary'
assert_forecast(forecast, forecast_type = NULL, verbose = TRUE, ...)
## S3 method for class 'forecast_point'
assert_forecast(forecast, forecast_type = NULL, verbose = TRUE, ...)
## S3 method for class 'forecast_quantile'
assert_forecast(forecast, forecast_type = NULL, verbose = TRUE, ...)
## S3 method for class 'forecast_sample'
assert_forecast(forecast, forecast_type = NULL, verbose = TRUE, ...)
assert_forecast(forecast, forecast_type = NULL, verbose = TRUE, ...)
## Default S3 method:
assert_forecast(forecast, forecast_type = NULL, verbose = TRUE, ...)
Arguments
forecast |
A forecast object (a validated data.table with predicted and observed values). |
forecast_type |
(optional) The forecast type you expect the forecasts
to have. If the forecast type as determined by |
verbose |
Logical. If |
... |
Currently unused. You cannot pass additional arguments to scoring
functions via |
Value
Returns NULL invisibly.
Examples
forecast <- as_forecast_binary(example_binary)
assert_forecast(forecast)
Validation common to all forecast types
Description
The function runs input checks that apply to all input data, regardless of forecast type. The function
asserts that the forecast is a data.table which has columns
observedandpredictedchecks the forecast type and forecast unit
checks there are no duplicate forecasts
if appropriate, checks the number of samples / quantiles is the same for all forecasts.
Usage
assert_forecast_generic(data, verbose = TRUE)
Arguments
data |
A data.table with forecasts and observed values that should be validated. |
verbose |
Logical. If |
Value
returns the input
Assert that forecast type is as expected
Description
Assert that forecast type is as expected
Usage
assert_forecast_type(data, actual = get_forecast_type(data), desired = NULL)
Arguments
data |
A forecast object. |
actual |
The actual forecast type of the data |
desired |
The desired forecast type of the data |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Assert that inputs are correct for binary forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring binary forecasts.
Usage
assert_input_binary(observed, predicted)
Arguments
observed |
Input to be checked. Should be a factor of length n with
exactly two levels, holding the observed values.
The highest factor level is assumed to be the reference level. This means
that |
predicted |
Input to be checked. |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Assert that inputs are correct for categorical forecasts
Description
Function assesses whether the inputs correspond to the requirements for scoring categorical, i.e. either nominal or ordinal forecasts.
Usage
assert_input_categorical(observed, predicted, predicted_label, ordered = NA)
Arguments
observed |
Input to be checked. Should be a factor of length n with N levels holding the observed values. n is the number of observations and N is the number of possible outcomes the observed values can assume. |
predicted |
Input to be checked. Should be nxN matrix of predicted
probabilities, n (number of rows) being the number of data points and N
(number of columns) the number of possible outcomes the observed values
can assume.
If |
predicted_label |
Factor of length N with N levels, where N is the number of possible outcomes the observed values can assume. |
ordered |
Value indicating whether factors have to be ordered or not.
Defaults to |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Assert that inputs are correct for interval-based forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring interval-based forecasts.
Usage
assert_input_interval(observed, lower, upper, interval_range)
Arguments
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
lower |
Input to be checked. Should be a numeric vector of size n that holds the predicted value for the lower bounds of the prediction intervals. |
upper |
Input to be checked. Should be a numeric vector of size n that holds the predicted value for the upper bounds of the prediction intervals. |
interval_range |
Input to be checked. Should be a vector of size n that denotes the interval range in percent. E.g. a value of 50 denotes a (25%, 75%) prediction interval. |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Assert that inputs are correct for nominal forecasts
Description
Function assesses whether the inputs correspond to the requirements for scoring nominal forecasts.
Usage
assert_input_nominal(observed, predicted, predicted_label)
Arguments
observed |
Input to be checked. Should be an unordered factor of length n with N levels holding the observed values. n is the number of observations and N is the number of possible outcomes the observed values can assume. |
predicted |
Input to be checked. Should be nxN matrix of predicted
probabilities, n (number of rows) being the number of data points and N
(number of columns) the number of possible outcomes the observed values
can assume.
If |
predicted_label |
Unordered factor of length N with N levels, where N is the number of possible outcomes the observed values can assume. |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Assert that inputs are correct for ordinal forecasts
Description
Function assesses whether the inputs correspond to the requirements for scoring ordinal forecasts.
Usage
assert_input_ordinal(observed, predicted, predicted_label)
Arguments
observed |
Input to be checked. Should be an ordered factor of length n with N levels holding the observed values. n is the number of observations and N is the number of possible outcomes the observed values can assume. |
predicted |
Input to be checked. Should be nxN matrix of predicted
probabilities, n (number of rows) being the number of data points and N
(number of columns) the number of possible outcomes the observed values
can assume.
If |
predicted_label |
Ordered factor of length N with N levels, where N is the number of possible outcomes the observed values can assume. |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Assert that inputs are correct for point forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring point forecasts.
Usage
assert_input_point(observed, predicted)
Arguments
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
predicted |
Input to be checked. Should be a numeric vector with the predicted values of size n. |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Assert that inputs are correct for quantile-based forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring quantile-based forecasts.
Usage
assert_input_quantile(
observed,
predicted,
quantile_level,
unique_quantile_levels = TRUE
)
Arguments
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
predicted |
Input to be checked. Should be nxN matrix of predictive
quantiles, n (number of rows) being the number of data points and N
(number of columns) the number of quantiles per forecast.
If |
quantile_level |
Input to be checked. Should be a vector of size N that denotes the quantile levels corresponding to the columns of the prediction matrix. |
unique_quantile_levels |
Whether the quantile levels are required to be
unique ( |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Assert that inputs are correct for sample-based forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring sample-based forecasts.
Usage
assert_input_sample(observed, predicted)
Arguments
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
predicted |
Input to be checked. Should be a numeric nxN matrix of
predictive samples, n (number of rows) being the number of data points and
N (number of columns) the number of samples per forecast.
If |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Validate an object of class scores
Description
This function validates an object of class scores, checking
that it has the correct class and that it has a metrics attribute.
Usage
assert_scores(scores)
Arguments
scores |
A data.table or similar with scores as produced by |
Value
Returns NULL invisibly
Determines bias of quantile forecasts
Description
Determines bias from quantile forecasts. For an increasing number of quantiles this measure converges against the sample based bias version for integer and continuous forecasts.
Usage
bias_quantile(observed, predicted, quantile_level, na.rm = TRUE)
Arguments
observed |
Numeric vector of size n with the observed values. |
predicted |
Numeric nxN matrix of predictive
quantiles, n (number of rows) being the number of forecasts (corresponding
to the number of observed values) and N
(number of columns) the number of quantiles per forecast.
If |
quantile_level |
Vector of of size N with the quantile levels for which predictions were made. Note that if this does not contain the median (0.5) then the median is imputed as being the mean of the two innermost quantiles. |
na.rm |
Logical. Should missing values be removed? |
Details
For quantile forecasts, bias is measured as
B_t = (1 - 2 \cdot \max \{i | q_{t,i} \in Q_t \land q_{t,i} \leq x_t\})
\mathbf{1}( x_t \leq q_{t, 0.5}) \\
+ (1 - 2 \cdot \min \{i | q_{t,i} \in Q_t \land q_{t,i} \geq x_t\})
1( x_t \geq q_{t, 0.5}),
where Q_t is the set of quantiles that form the predictive
distribution at time t and x_t is the observed value. For
consistency, we define Q_t such that it always includes the element
q_{t, 0} = - \infty and q_{t,1} = \infty.
1() is the indicator function that is 1 if the
condition is satisfied and 0 otherwise.
In clearer terms, bias B_t is:
-
1 - 2 \cdotthe maximum percentile rank for which the corresponding quantile is still smaller than or equal to the observed value, if the observed value is smaller than the median of the predictive distribution. -
1 - 2 \cdotthe minimum percentile rank for which the corresponding quantile is still larger than or equal to the observed value if the observed value is larger than the median of the predictive distribution.. -
0if the observed value is exactly the median (both terms cancel out)
Bias can assume values between -1 and 1 and is 0 ideally (i.e. unbiased).
Note that if the given quantiles do not contain the median, the median is imputed as a linear interpolation of the two innermost quantiles. If the median is not available and cannot be imputed, an error will be thrown. Note that in order to compute bias, quantiles must be non-decreasing with increasing quantile levels.
For a large enough number of quantiles, the
percentile rank will equal the proportion of predictive samples below the
observed value, and the bias metric coincides with the one for
continuous forecasts (see bias_sample()).
Value
scalar with the quantile bias for a single quantile prediction
Input format

Overview of required input format for quantile-based forecasts
Examples
predicted <- matrix(c(1.5:23.5, 3.3:25.3), nrow = 2, byrow = TRUE)
quantile_level <- c(0.01, 0.025, seq(0.05, 0.95, 0.05), 0.975, 0.99)
observed <- c(15, 12.4)
bias_quantile(observed, predicted, quantile_level)
Compute bias for a single vector of quantile predictions
Description
Internal function to compute bias for a single observed value, a vector of predicted values and a vector of quantiles.
Usage
bias_quantile_single_vector(observed, predicted, quantile_level, na.rm)
Arguments
observed |
Scalar with the observed value. |
predicted |
Vector of length N (corresponding to the number of quantiles) that holds predictions. |
quantile_level |
Vector of of size N with the quantile levels for which predictions were made. Note that if this does not contain the median (0.5) then the median is imputed as being the mean of the two innermost quantiles. |
na.rm |
Logical. Should missing values be removed? |
Value
scalar with the quantile bias for a single quantile prediction
Determine bias of forecasts
Description
Determines bias from predictive Monte-Carlo samples. The function automatically recognises whether forecasts are continuous or integer valued and adapts the Bias function accordingly.
Usage
bias_sample(observed, predicted)
Arguments
observed |
A vector with observed values of size n |
predicted |
nxN matrix of predictive samples, n (number of rows) being
the number of data points and N (number of columns) the number of Monte
Carlo samples. Alternatively, |
Details
For continuous forecasts, Bias is measured as
B_t (P_t, x_t) = 1 - 2 * (P_t (x_t))
where P_t is the empirical cumulative distribution function of the
prediction for the observed value x_t. Computationally, P_t (x_t) is
just calculated as the fraction of predictive samples for x_t
that are smaller than x_t.
For integer valued forecasts, Bias is measured as
B_t (P_t, x_t) = 1 - (P_t (x_t) + P_t (x_t + 1))
to adjust for the integer nature of the forecasts.
In both cases, Bias can assume values between -1 and 1 and is 0 ideally.
Value
Numeric vector of length n with the biases of the predictive samples with respect to the observed values.
Input format

Overview of required input format for sample-based forecasts
References
The integer valued Bias function is discussed in Assessing the performance of real-time epidemic forecasts: A case study of Ebola in the Western Area region of Sierra Leone, 2014-15 Funk S, Camacho A, Kucharski AJ, Lowe R, Eggo RM, et al. (2019) Assessing the performance of real-time epidemic forecasts: A case study of Ebola in the Western Area region of Sierra Leone, 2014-15. PLOS Computational Biology 15(2): e1006785. doi:10.1371/journal.pcbi.1006785
Examples
## integer valued forecasts
observed <- rpois(30, lambda = 1:30)
predicted <- replicate(200, rpois(n = 30, lambda = 1:30))
bias_sample(observed, predicted)
## continuous forecasts
observed <- rnorm(30, mean = 1:30)
predicted <- replicate(200, rnorm(30, mean = 1:30))
bias_sample(observed, predicted)
Check column names are present in a data.frame
Description
The functions loops over the column names and checks whether they are present. If an issue is encountered, the function immediately stops and returns a message with the first issue encountered.
Usage
check_columns_present(data, columns)
Arguments
data |
A data.frame or similar to be checked |
columns |
A character vector of column names to check |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Check Inputs Have Matching Dimensions
Description
Function assesses whether input dimensions match. In the following, n is the number of observations / forecasts. Scalar values may be repeated to match the length of the other input. Allowed options are therefore:
-
observedis vector of length 1 or length n -
predictedis:a vector of of length 1 or length n
a matrix with n rows and 1 column
Usage
check_dims_ok_point(observed, predicted)
Arguments
observed |
Input to be checked. Should be a factor of length n with
exactly two levels, holding the observed values.
The highest factor level is assumed to be the reference level. This means
that |
predicted |
Input to be checked. |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Check that there are no duplicate forecasts
Description
Runs get_duplicate_forecasts() and returns a message if an issue is
encountered
Usage
check_duplicates(data)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Check that inputs are correct for binary forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring binary forecasts.
Usage
check_input_binary(observed, predicted)
Arguments
observed |
Input to be checked. Should be a factor of length n with
exactly two levels, holding the observed values.
The highest factor level is assumed to be the reference level. This means
that |
predicted |
Input to be checked. |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Check that inputs are correct for interval-based forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring interval-based forecasts.
Usage
check_input_interval(observed, lower, upper, interval_range)
Arguments
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
lower |
Input to be checked. Should be a numeric vector of size n that holds the predicted value for the lower bounds of the prediction intervals. |
upper |
Input to be checked. Should be a numeric vector of size n that holds the predicted value for the upper bounds of the prediction intervals. |
interval_range |
Input to be checked. Should be a vector of size n that denotes the interval range in percent. E.g. a value of 50 denotes a (25%, 75%) prediction interval. |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Check that inputs are correct for point forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring point forecasts.
Usage
check_input_point(observed, predicted)
Arguments
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
predicted |
Input to be checked. Should be a numeric vector with the predicted values of size n. |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Check that inputs are correct for quantile-based forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring quantile-based forecasts.
Usage
check_input_quantile(observed, predicted, quantile_level)
Arguments
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
predicted |
Input to be checked. Should be nxN matrix of predictive
quantiles, n (number of rows) being the number of data points and N
(number of columns) the number of quantiles per forecast.
If |
quantile_level |
Input to be checked. Should be a vector of size N that denotes the quantile levels corresponding to the columns of the prediction matrix. |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Check that inputs are correct for sample-based forecast
Description
Function assesses whether the inputs correspond to the requirements for scoring sample-based forecasts.
Usage
check_input_sample(observed, predicted)
Arguments
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
predicted |
Input to be checked. Should be a numeric nxN matrix of
predictive samples, n (number of rows) being the number of data points and
N (number of columns) the number of samples per forecast.
If |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Check that all forecasts have the same number of rows
Description
Helper function that checks the number of rows (corresponding e.g to quantiles or samples) per forecast. If the number of quantiles or samples is the same for all forecasts, it returns TRUE and a string with an error message otherwise.
Usage
check_number_per_forecast(data, forecast_unit)
Arguments
data |
A data.frame or similar to be checked |
forecast_unit |
Character vector denoting the unit of a single forecast. |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Check whether an input is an atomic vector of mode 'numeric'
Description
Helper function to check whether an input is a numeric vector.
Usage
check_numeric_vector(x, ...)
Arguments
x |
input to check |
... |
Arguments passed on to
|
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Helper function to convert assert statements into checks
Description
Tries to execute an expression. Internally, this is used to
see whether assertions fail when checking inputs (i.e. to convert an
assert_*() statement into a check). If the expression fails, the error
message is returned. If the expression succeeds, TRUE is returned.
Usage
check_try(expr)
Arguments
expr |
an expression to be evaluated |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Clean forecast object
Description
The function makes it possible to silently validate an object. In addition, it can return a copy of the data and remove rows with missing values.
Usage
clean_forecast(forecast, copy = FALSE, na.omit = FALSE)
Arguments
forecast |
A forecast object (a validated data.table with predicted and observed values). |
copy |
Logical, default is |
na.omit |
Logical, default is |
Compare a subset of common forecasts
Description
This function compares two comparators based on the subset of forecasts for which
both comparators have made a prediction. It gets called
from pairwise_comparison_one_group(), which handles the
comparison of multiple comparators on a single set of forecasts (there are no
subsets of forecasts to be distinguished). pairwise_comparison_one_group()
in turn gets called from from get_pairwise_comparisons() which can handle
pairwise comparisons for a set of forecasts with multiple subsets, e.g.
pairwise comparisons for one set of forecasts, but done separately for two
different forecast targets.
Usage
compare_forecasts(
scores,
compare = "model",
name_comparator1,
name_comparator2,
metric,
one_sided = FALSE,
test_type = c("non_parametric", "permutation", NULL),
n_permutations = 999
)
Arguments
scores |
An object of class |
compare |
Character vector with a single colum name that defines the elements for the pairwise comparison. For example, if this is set to "model" (the default), then elements of the "model" column will be compared. |
name_comparator1 |
Character, name of the first comparator |
name_comparator2 |
Character, name of the comparator to compare against |
metric |
A string with the name of the metric for which a relative skill shall be computed. By default this is either "crps", "wis" or "brier_score" if any of these are available. |
one_sided |
Boolean, default is |
test_type |
Character, either "non_parametric" (the default), "permutation", or NULL. This determines which kind of test shall be conducted to determine p-values. If NULL, no test will be conducted and p-values will be NA. |
n_permutations |
Numeric, the number of permutations for a permutation test. Default is 999. |
Value
A list with mean score ratios and p-values for the comparison between two comparators
Author(s)
Johannes Bracher, johannes.bracher@kit.edu
Nikos Bosse nikosbosse@gmail.com
(Continuous) ranked probability score
Description
Wrapper around the crps_sample()
function from the
scoringRules package. Can be used for continuous as well as integer
valued forecasts
The Continuous ranked probability score (CRPS) can be interpreted as the sum of three components: overprediction, underprediction and dispersion. "Dispersion" is defined as the CRPS of the median forecast $m$. If an observation $y$ is greater than $m$ then overprediction is defined as the CRPS of the forecast for $y$ minus the dispersion component, and underprediction is zero. If, on the other hand, $y<m$ then underprediction is defined as the CRPS of the forecast for $y$ minus the dispersion component, and overprediction is zero.
The overprediction, underprediction and dispersion components correspond to
those of the wis().
Usage
crps_sample(observed, predicted, separate_results = FALSE, ...)
dispersion_sample(observed, predicted, ...)
overprediction_sample(observed, predicted, ...)
underprediction_sample(observed, predicted, ...)
Arguments
observed |
A vector with observed values of size n |
predicted |
nxN matrix of predictive samples, n (number of rows) being
the number of data points and N (number of columns) the number of Monte
Carlo samples. Alternatively, |
separate_results |
Logical. If |
... |
Additional arguments passed on to |
Value
Vector with scores.
dispersion_sample(): a numeric vector with dispersion values (one per
observation).
overprediction_sample(): a numeric vector with overprediction values
(one per observation).
underprediction_sample(): a numeric vector with underprediction values (one per
observation).
Input format

Overview of required input format for sample-based forecasts
References
Alexander Jordan, Fabian Krüger, Sebastian Lerch, Evaluating Probabilistic Forecasts with scoringRules, https://www.jstatsoft.org/article/view/v090i12
Examples
observed <- rpois(30, lambda = 1:30)
predicted <- replicate(200, rpois(n = 30, lambda = 1:30))
crps_sample(observed, predicted)
Documentation template for assert functions
Description
Documentation template for assert functions
Arguments
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
Value
Returns NULL invisibly if the assertion was successful and throws an error otherwise.
Documentation template for check functions
Description
Documentation template for check functions
Arguments
data |
A data.frame or similar to be checked |
observed |
Input to be checked. Should be a numeric vector with the observed values of size n. |
columns |
A character vector of column names to check |
Value
Returns TRUE if the check was successful and a string with an error message otherwise.
Documentation template for test functions
Description
Documentation template for test functions
Value
Returns TRUE if the check was successful and FALSE otherwise
Dawid-Sebastiani score
Description
Wrapper around the dss_sample()
function from the
scoringRules package.
Usage
dss_sample(observed, predicted, ...)
Arguments
observed |
A vector with observed values of size n |
predicted |
nxN matrix of predictive samples, n (number of rows) being
the number of data points and N (number of columns) the number of Monte
Carlo samples. Alternatively, |
... |
Additional arguments passed to dss_sample() from the scoringRules package. |
Value
Vector with scores.
Input format

Overview of required input format for sample-based forecasts
References
Alexander Jordan, Fabian Krüger, Sebastian Lerch, Evaluating Probabilistic Forecasts with scoringRules, https://www.jstatsoft.org/article/view/v090i12
Examples
observed <- rpois(30, lambda = 1:30)
predicted <- replicate(200, rpois(n = 30, lambda = 1:30))
dss_sample(observed, predicted)
Ensure that an object is a data.table
Description
This function ensures that an object is a data table.
If the object is not a data table, it is converted to one. If the object
is a data table, a copy of the object is returned.
Usage
ensure_data.table(data)
Arguments
data |
An object to ensure is a data table. |
Value
A data.table/a copy of an existing data.table.
Binary forecast example data
Description
A data set with binary predictions for COVID-19 cases and deaths constructed from data submitted to the European Forecast Hub.
Usage
example_binary
Format
An object of class forecast_binary (see as_forecast_binary())
with the following columns:
- location
the country for which a prediction was made
- location_name
name of the country for which a prediction was made
- target_end_date
the date for which a prediction was made
- target_type
the target to be predicted (cases or deaths)
- observed
A factor with observed values
- forecast_date
the date on which a prediction was made
- model
name of the model that generated the forecasts
- horizon
forecast horizon in weeks
- predicted
predicted value
Details
Predictions in the data set were constructed based on the continuous example data by looking at the number of samples below the mean prediction. The outcome was constructed as whether or not the actually observed value was below or above that mean prediction. This should not be understood as sound statistical practice, but rather as a practical way to create an example data set.
The data was created using the script create-example-data.R in the inst/ folder (or the top level folder in a compiled package).
Source
Nominal example data
Description
A data set with predictions for COVID-19 cases and deaths submitted to the European Forecast Hub.
Usage
example_nominal
Format
An object of class forecast_nominal
(see as_forecast_nominal()) with the following columns:
- location
the country for which a prediction was made
- target_end_date
the date for which a prediction was made
- target_type
the target to be predicted (cases or deaths)
- observed
Numeric: observed values
- location_name
name of the country for which a prediction was made
- forecast_date
the date on which a prediction was made
- predicted_label
outcome that a probabilty corresponds to
- predicted
predicted value
- model
name of the model that generated the forecasts
- horizon
forecast horizon in weeks
Details
The data was created using the script create-example-data.R in the inst/ folder (or the top level folder in a compiled package).
Source
Ordinal example data
Description
A data set with predictions for COVID-19 cases and deaths submitted to the European Forecast Hub.
Usage
example_ordinal
Format
An object of class forecast_ordinal
(see as_forecast_ordinal()) with the following columns:
- location
the country for which a prediction was made
- target_end_date
the date for which a prediction was made
- target_type
the target to be predicted (cases or deaths)
- observed
Numeric: observed values
- location_name
name of the country for which a prediction was made
- forecast_date
the date on which a prediction was made
- predicted_label
outcome that a probabilty corresponds to
- predicted
predicted value
- model
name of the model that generated the forecasts
- horizon
forecast horizon in weeks
Details
The data was created using the script create-example-data.R in the inst/ folder (or the top level folder in a compiled package).
Source
Point forecast example data
Description
A data set with predictions for COVID-19 cases and deaths submitted to the European Forecast Hub. This data set is like the quantile example data, only that the median has been replaced by a point forecast.
Usage
example_point
Format
An object of class forecast_point (see as_forecast_point())
with the following columns:
- location
the country for which a prediction was made
- target_end_date
the date for which a prediction was made
- target_type
the target to be predicted (cases or deaths)
- observed
observed values
- location_name
name of the country for which a prediction was made
- forecast_date
the date on which a prediction was made
- predicted
predicted value
- model
name of the model that generated the forecasts
- horizon
forecast horizon in weeks
Details
The data was created using the script create-example-data.R in the inst/ folder (or the top level folder in a compiled package).
Source
Quantile example data
Description
A data set with predictions for COVID-19 cases and deaths submitted to the European Forecast Hub.
Usage
example_quantile
Format
An object of class forecast_quantile
(see as_forecast_quantile()) with the following columns:
- location
the country for which a prediction was made
- target_end_date
the date for which a prediction was made
- target_type
the target to be predicted (cases or deaths)
- observed
Numeric: observed values
- location_name
name of the country for which a prediction was made
- forecast_date
the date on which a prediction was made
- quantile_level
quantile level of the corresponding prediction
- predicted
predicted value
- model
name of the model that generated the forecasts
- horizon
forecast horizon in weeks
Details
The data was created using the script create-example-data.R in the inst/ folder (or the top level folder in a compiled package).
Source
Continuous forecast example data
Description
A data set with continuous predictions for COVID-19 cases and deaths constructed from data submitted to the European Forecast Hub.
Usage
example_sample_continuous
Format
An object of class forecast_sample (see as_forecast_sample())
with the following columns:
- location
the country for which a prediction was made
- target_end_date
the date for which a prediction was made
- target_type
the target to be predicted (cases or deaths)
- observed
observed values
- location_name
name of the country for which a prediction was made
- forecast_date
the date on which a prediction was made
- model
name of the model that generated the forecasts
- horizon
forecast horizon in weeks
- predicted
predicted value
- sample_id
id for the corresponding sample
Details
The data was created using the script create-example-data.R in the inst/ folder (or the top level folder in a compiled package).
Source
Discrete forecast example data
Description
A data set with integer predictions for COVID-19 cases and deaths constructed from data submitted to the European Forecast Hub.
Usage
example_sample_discrete
Format
An object of class forecast_sample (see as_forecast_sample())
with the following columns:
- location
the country for which a prediction was made
- target_end_date
the date for which a prediction was made
- target_type
the target to be predicted (cases or deaths)
- observed
observed values
- location_name
name of the country for which a prediction was made
- forecast_date
the date on which a prediction was made
- model
name of the model that generated the forecasts
- horizon
forecast horizon in weeks
- predicted
predicted value
- sample_id
id for the corresponding sample
Details
The data was created using the script create-example-data.R in the inst/ folder (or the top level folder in a compiled package).
Source
Documentation template for forecast types
Description
Documentation template for forecast types
Forecast unit
In order to score forecasts, scoringutils needs to know which of the rows
of the data belong together and jointly form a single forecasts. This is
easy e.g. for point forecast, where there is one row per forecast. For
quantile or sample-based forecasts, however, there are multiple rows that
belong to a single forecast.
The forecast unit or unit of a single forecast is then described by the
combination of columns that uniquely identify a single forecast.
For example, we could have forecasts made by different models in various
locations at different time points, each for several weeks into the future.
The forecast unit could then be described as
forecast_unit = c("model", "location", "forecast_date", "forecast_horizon").
scoringutils automatically tries to determine the unit of a single
forecast. It uses all existing columns for this, which means that no columns
must be present that are unrelated to the forecast unit. As a very simplistic
example, if you had an additional row, "even", that is one if the row number
is even and zero otherwise, then this would mess up scoring as scoringutils
then thinks that this column was relevant in defining the forecast unit.
In order to avoid issues, we recommend setting the forecast unit explicitly,
using the forecast_unit argument. This will simply drop unneeded columns,
while making sure that all necessary, 'protected columns' like "predicted"
or "observed" are retained.
Calculate geometric mean
Description
Calculate geometric mean
Usage
geometric_mean(x)
Arguments
x |
Numeric vector of values for which to calculate the geometric mean. |
Details
Used in get_pairwise_comparisons().
Value
The geometric mean of the values in x. NA values are ignored.
Calculate correlation between metrics
Description
Calculate the correlation between different metrics for a data.frame of
scores as produced by score().
Usage
get_correlations(scores, metrics = get_metrics.scores(scores), ...)
Arguments
scores |
An object of class |
metrics |
A character vector with the metrics to show. If set to
|
... |
Additional arguments to pass down to |
Value
An object of class scores (a data.table with an additional
attribute metrics holding the names of the scores) with correlations
between different metrics
Examples
library(magrittr) # pipe operator
scores <- example_quantile %>%
as_forecast_quantile() %>%
score()
get_correlations(scores)
Get quantile and interval coverage values for quantile-based forecasts
Description
For a validated forecast object in a quantile-based format
(see as_forecast_quantile() for more information), this function computes:
interval coverage of central prediction intervals
quantile coverage for predictive quantiles
the deviation between desired and actual coverage (both for interval and quantile coverage)
Coverage values are computed for a specific level of grouping, as specified
in the by argument. By default, coverage values are computed per model.
Interval coverage
Interval coverage for a given interval range is defined as the proportion of observations that fall within the corresponding central prediction intervals. Central prediction intervals are symmetric around the median and formed by two quantiles that denote the lower and upper bound. For example, the 50% central prediction interval is the interval between the 0.25 and 0.75 quantiles of the predictive distribution.
Quantile coverage
Quantile coverage for a given quantile level is defined as the proportion of
observed values that are smaller than the corresponding predictive quantile.
For example, the 0.5 quantile coverage is the proportion of observed values
that are smaller than the 0.5 quantile of the predictive distribution.
Just as above, for a single observation and the quantile of a single
predictive distribution, the value will either be TRUE or FALSE.
Coverage deviation
The coverage deviation is the difference between the desired coverage (can be either interval or quantile coverage) and the actual coverage. For example, if the desired coverage is 90% and the actual coverage is 80%, the coverage deviation is -0.1.
Usage
get_coverage(forecast, by = "model")
Arguments
forecast |
A forecast object (a validated data.table with predicted and observed values). |
by |
character vector that denotes the level of grouping for which the
coverage values should be computed. By default ( |
Value
A data.table with columns as specified in by and additional
columns for the coverage values described above
a data.table with columns "interval_coverage",
"interval_coverage_deviation", "quantile_coverage",
"quantile_coverage_deviation" and the columns specified in by.
Examples
library(magrittr) # pipe operator
example_quantile %>%
as_forecast_quantile() %>%
get_coverage(by = "model")
Find duplicate forecasts
Description
Internal helper function to identify duplicate forecasts, i.e. instances where there is more than one forecast for the same prediction target.
Usage
get_duplicate_forecasts(data, forecast_unit = NULL, counts = FALSE)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
forecast_unit |
(optional) Name of the columns in |
counts |
Should the output show the number of duplicates per forecast
unit instead of the individual duplicated rows? Default is |
Value
A data.frame with all rows for which a duplicate forecast was found
Examples
example <- rbind(example_quantile, example_quantile[1000:1010])
get_duplicate_forecasts(example)
Count number of available forecasts
Description
Given a data set with forecasts, this function counts the number of
available forecasts.
The level of grouping can be specified using the by argument (e.g. to
count the number of forecasts per model, or the number of forecasts per
model and location).
This is useful to determine whether there are any missing forecasts.
Usage
get_forecast_counts(
forecast,
by = get_forecast_unit(forecast),
collapse = c("quantile_level", "sample_id")
)
Arguments
forecast |
A forecast object (a validated data.table with predicted and observed values). |
by |
character vector or |
collapse |
character vector (default: |
Value
A data.table with columns as specified in by and an additional
column "count" with the number of forecasts.
Examples
library(magrittr) # pipe operator
example_quantile %>%
as_forecast_quantile() %>%
get_forecast_counts(by = c("model", "target_type"))
Get forecast type from forecast object
Description
Get forecast type from forecast object
Usage
get_forecast_type(forecast)
Arguments
forecast |
A forecast object (a validated data.table with predicted and observed values). |
Value
Character vector of length one with the forecast type.
Get unit of a single forecast
Description
Helper function to get the unit of a single forecast, i.e.
the column names that define where a single forecast was made for.
This just takes all columns that are available in the data and subtracts
the columns that are protected, i.e. those returned by
get_protected_columns() as well as the names of the metrics that were
specified during scoring, if any.
Usage
get_forecast_unit(data)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
Value
A character vector with the column names that define the unit of a single forecast
Forecast unit
In order to score forecasts, scoringutils needs to know which of the rows
of the data belong together and jointly form a single forecasts. This is
easy e.g. for point forecast, where there is one row per forecast. For
quantile or sample-based forecasts, however, there are multiple rows that
belong to a single forecast.
The forecast unit or unit of a single forecast is then described by the
combination of columns that uniquely identify a single forecast.
For example, we could have forecasts made by different models in various
locations at different time points, each for several weeks into the future.
The forecast unit could then be described as
forecast_unit = c("model", "location", "forecast_date", "forecast_horizon").
scoringutils automatically tries to determine the unit of a single
forecast. It uses all existing columns for this, which means that no columns
must be present that are unrelated to the forecast unit. As a very simplistic
example, if you had an additional row, "even", that is one if the row number
is even and zero otherwise, then this would mess up scoring as scoringutils
then thinks that this column was relevant in defining the forecast unit.
In order to avoid issues, we recommend setting the forecast unit explicitly,
using the forecast_unit argument. This will simply drop unneeded columns,
while making sure that all necessary, 'protected columns' like "predicted"
or "observed" are retained.
Get metrics
Description
Generic function to to obtain default metrics available for scoring or metrics that were used for scoring.
If called on a
forecastobject it returns a list of functions that can be used for scoring.If called on a
scoresobject (seescore()), it returns a character vector with the names of the metrics that were used for scoring.
See the documentation for the actual methods in the See Also section below
for more details. Alternatively call ?get_metrics.<forecast_type> or
?get_metrics.scores.
Usage
get_metrics(x, ...)
Arguments
x |
A |
... |
Additional arguments passed to the method. |
See Also
Other get_metrics functions:
get_metrics.forecast_binary(),
get_metrics.forecast_nominal(),
get_metrics.forecast_ordinal(),
get_metrics.forecast_point(),
get_metrics.forecast_quantile(),
get_metrics.forecast_sample(),
get_metrics.scores()
Get default metrics for binary forecasts
Description
For binary forecasts, the default scoring rules are:
"brier_score" =
brier_score()"log_score" =
logs_binary()
Usage
## S3 method for class 'forecast_binary'
get_metrics(x, select = NULL, exclude = NULL, ...)
Arguments
x |
A forecast object (a validated data.table with predicted and
observed values, see |
select |
A character vector of scoring rules to select from the list. If
|
exclude |
A character vector of scoring rules to exclude from the list.
If |
... |
unused |
Value
A list of scoring functions.
Input format
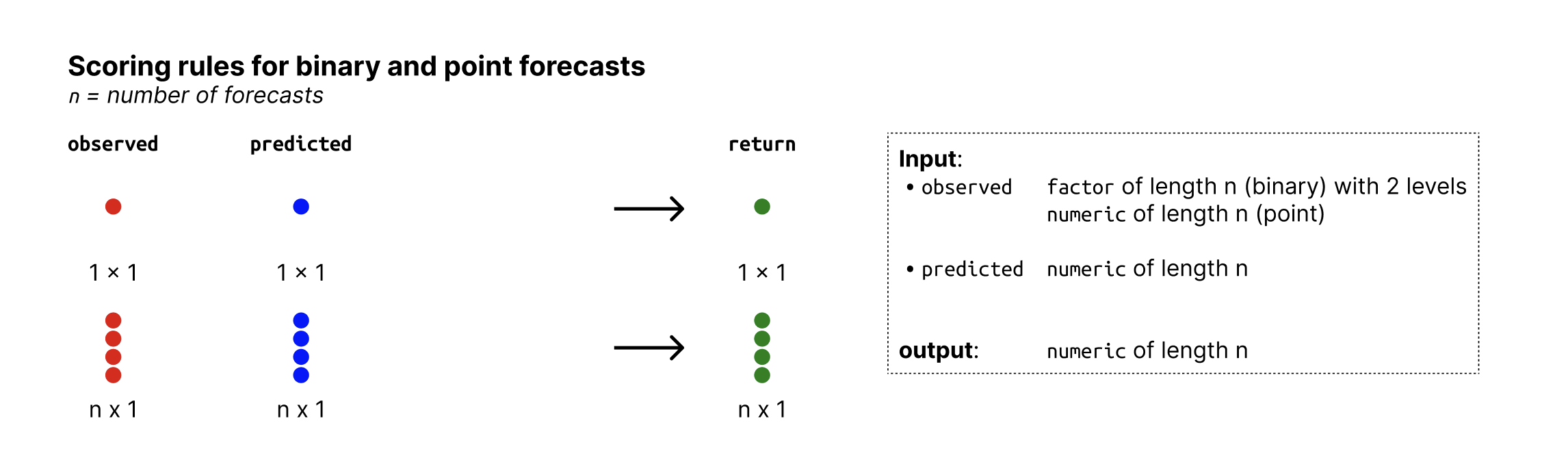
Overview of required input format for binary and point forecasts
See Also
Other get_metrics functions:
get_metrics(),
get_metrics.forecast_nominal(),
get_metrics.forecast_ordinal(),
get_metrics.forecast_point(),
get_metrics.forecast_quantile(),
get_metrics.forecast_sample(),
get_metrics.scores()
Examples
get_metrics(example_binary)
get_metrics(example_binary, select = "brier_score")
get_metrics(example_binary, exclude = "log_score")
Get default metrics for nominal forecasts
Description
For nominal forecasts, the default scoring rule is:
"log_score" =
logs_categorical()
Usage
## S3 method for class 'forecast_nominal'
get_metrics(x, select = NULL, exclude = NULL, ...)
Arguments
x |
A forecast object (a validated data.table with predicted and
observed values, see |
select |
A character vector of scoring rules to select from the list. If
|
exclude |
A character vector of scoring rules to exclude from the list.
If |
... |
unused |
See Also
Other get_metrics functions:
get_metrics(),
get_metrics.forecast_binary(),
get_metrics.forecast_ordinal(),
get_metrics.forecast_point(),
get_metrics.forecast_quantile(),
get_metrics.forecast_sample(),
get_metrics.scores()
Examples
get_metrics(example_nominal)
Get default metrics for nominal forecasts
Description
For ordinal forecasts, the default scoring rules are:
"log_score" =
logs_categorical()"rps" =
rps_ordinal()
Usage
## S3 method for class 'forecast_ordinal'
get_metrics(x, select = NULL, exclude = NULL, ...)
Arguments
x |
A forecast object (a validated data.table with predicted and
observed values, see |
select |
A character vector of scoring rules to select from the list. If
|
exclude |
A character vector of scoring rules to exclude from the list.
If |
... |
unused |
See Also
Other get_metrics functions:
get_metrics(),
get_metrics.forecast_binary(),
get_metrics.forecast_nominal(),
get_metrics.forecast_point(),
get_metrics.forecast_quantile(),
get_metrics.forecast_sample(),
get_metrics.scores()
Examples
get_metrics(example_ordinal)
Get default metrics for point forecasts
Description
For point forecasts, the default scoring rules are:
A note of caution: Every scoring rule for a point forecast is implicitly minimised by a specific aspect of the predictive distribution (see Gneiting, 2011).
The mean squared error, for example, is only a meaningful scoring rule if the forecaster actually reported the mean of their predictive distribution as a point forecast. If the forecaster reported the median, then the mean absolute error would be the appropriate scoring rule. If the scoring rule and the predictive task do not align, the results will be misleading.
Failure to respect this correspondence can lead to grossly misleading results! Consider the example in the section below.
Usage
## S3 method for class 'forecast_point'
get_metrics(x, select = NULL, exclude = NULL, ...)
Arguments
x |
A forecast object (a validated data.table with predicted and
observed values, see |
select |
A character vector of scoring rules to select from the list. If
|
exclude |
A character vector of scoring rules to exclude from the list.
If |
... |
unused |
Input format

Overview of required input format for binary and point forecasts
References
Making and Evaluating Point Forecasts, Gneiting, Tilmann, 2011, Journal of the American Statistical Association.
See Also
Other get_metrics functions:
get_metrics(),
get_metrics.forecast_binary(),
get_metrics.forecast_nominal(),
get_metrics.forecast_ordinal(),
get_metrics.forecast_quantile(),
get_metrics.forecast_sample(),
get_metrics.scores()
Examples
get_metrics(example_point, select = "ape")
library(magrittr)
set.seed(123)
n <- 500
observed <- rnorm(n, 5, 4)^2
predicted_mu <- mean(observed)
predicted_not_mu <- predicted_mu - rnorm(n, 10, 2)
df <- data.frame(
model = rep(c("perfect", "bad"), each = n),
predicted = c(rep(predicted_mu, n), predicted_not_mu),
observed = rep(observed, 2),
id = rep(1:n, 2)
) %>%
as_forecast_point()
score(df) %>%
summarise_scores()
Get default metrics for quantile-based forecasts
Description
For quantile-based forecasts, the default scoring rules are:
"wis" =
wis()"overprediction" =
overprediction_quantile()"underprediction" =
underprediction_quantile()"dispersion" =
dispersion_quantile()"bias" =
bias_quantile()"interval_coverage_50" =
interval_coverage()"interval_coverage_90" = purrr::partial( interval_coverage, interval_range = 90 )
"ae_median" =
ae_median_quantile()
Note: The interval_coverage_90 scoring rule is created by modifying
interval_coverage(), making use of the function purrr::partial().
This construct allows the function to deal with arbitrary arguments in ...,
while making sure that only those that interval_coverage() can
accept get passed on to it. interval_range = 90 is set in the function
definition, as passing an argument interval_range = 90 to score() would
mean it would also get passed to interval_coverage_50.
Usage
## S3 method for class 'forecast_quantile'
get_metrics(x, select = NULL, exclude = NULL, ...)
Arguments
x |
A forecast object (a validated data.table with predicted and
observed values, see |
select |
A character vector of scoring rules to select from the list. If
|
exclude |
A character vector of scoring rules to exclude from the list.
If |
... |
unused |
Input format

Overview of required input format for quantile-based forecasts
See Also
Other get_metrics functions:
get_metrics(),
get_metrics.forecast_binary(),
get_metrics.forecast_nominal(),
get_metrics.forecast_ordinal(),
get_metrics.forecast_point(),
get_metrics.forecast_sample(),
get_metrics.scores()
Examples
get_metrics(example_quantile, select = "wis")
Get default metrics for sample-based forecasts
Description
For sample-based forecasts, the default scoring rules are:
"crps" =
crps_sample()"overprediction" =
overprediction_sample()"underprediction" =
underprediction_sample()"dispersion" =
dispersion_sample()"log_score" =
logs_sample()"dss" =
dss_sample()"mad" =
mad_sample()"bias" =
bias_sample()"ae_median" =
ae_median_sample()"se_mean" =
se_mean_sample()
Usage
## S3 method for class 'forecast_sample'
get_metrics(x, select = NULL, exclude = NULL, ...)
Arguments
x |
A forecast object (a validated data.table with predicted and
observed values, see |
select |
A character vector of scoring rules to select from the list. If
|
exclude |
A character vector of scoring rules to exclude from the list.
If |
... |
unused |
Input format

Overview of required input format for sample-based forecasts
See Also
Other get_metrics functions:
get_metrics(),
get_metrics.forecast_binary(),
get_metrics.forecast_nominal(),
get_metrics.forecast_ordinal(),
get_metrics.forecast_point(),
get_metrics.forecast_quantile(),
get_metrics.scores()
Examples
get_metrics(example_sample_continuous, exclude = "mad")
Get names of the metrics that were used for scoring
Description
When applying a scoring rule via score(), the names of the scoring rules
become column names of the
resulting data.table. In addition, an attribute metrics will be
added to the output, holding the names of the scores as a vector.
This is done so that functions like get_forecast_unit() or
summarise_scores() can still identify which columns are part of the
forecast unit and which hold a score.
get_metrics() accesses and returns the metrics attribute. If there is no
attribute, the function will return NULL (or, if error = TRUE will
produce an error instead). In addition, it checks the column names of the
input for consistency with the data stored in the metrics attribute.
Handling a missing or inconsistent metrics attribute:
If the metrics attribute is missing or is not consistent with the column names of the data.table, you can either
run
score()again, specifying names for the scoring rules manually, oradd/update the attribute manually using
attr(scores, "metrics") <- c("names", "of", "your", "scores")(the order does not matter).
Usage
## S3 method for class 'scores'
get_metrics(x, error = FALSE, ...)
Arguments
x |
A |
error |
Throw an error if there is no attribute called |
... |
unused |
Value
Character vector with the names of the scoring rules that were used for scoring.
See Also
Other get_metrics functions:
get_metrics(),
get_metrics.forecast_binary(),
get_metrics.forecast_nominal(),
get_metrics.forecast_ordinal(),
get_metrics.forecast_point(),
get_metrics.forecast_quantile(),
get_metrics.forecast_sample()
Obtain pairwise comparisons between models
Description
Compare scores obtained by different models in a pairwise tournament. All combinations of two models are compared against each other based on the overlapping set of available forecasts common to both models.
The input should be a scores object as produced by score(). Note that
adding additional unrelated columns can unpredictably change results, as
all present columns are taken into account when determining the set of
overlapping forecasts between two models.
The output of the pairwise comparisons is a set of mean score ratios, relative skill scores and p-values.
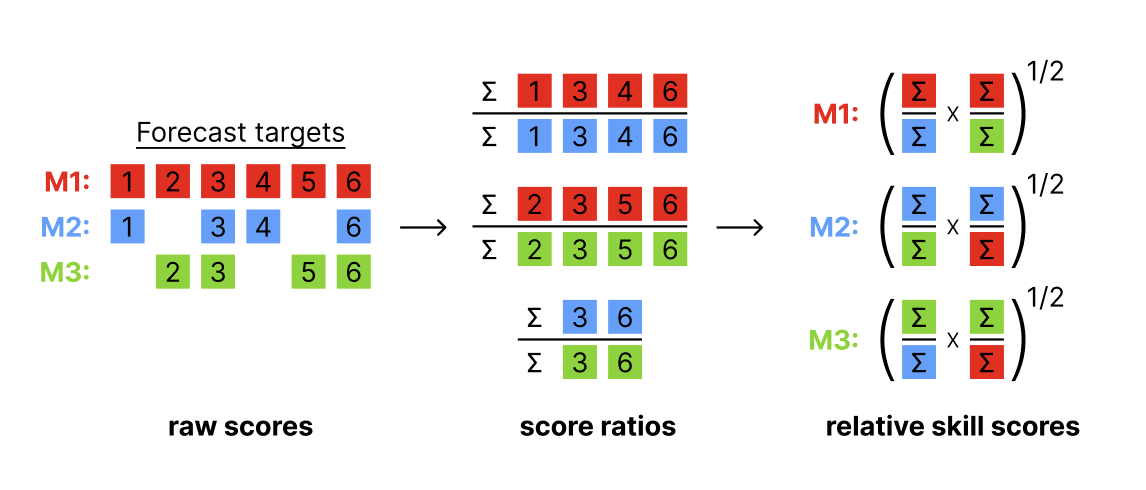
Illustration of the pairwise comparison process.
Mean score ratios
For every pair of two models, a mean score ratio is computed. This is simply the mean score of the first model divided by the mean score of the second. Mean score ratios are computed based on the set of overlapping forecasts between the two models. That means that only scores for those targets are taken into account for which both models have submitted a forecast.
(Scaled) Relative skill scores
The relative score of a model is the geometric mean of all mean score ratios which involve that model. If a baseline is provided, scaled relative skill scores will be calculated as well. Scaled relative skill scores are simply the relative skill score of a model divided by the relative skill score of the baseline model.
p-values
In addition, the function computes p-values for the comparison between two
models (again based on the set of overlapping forecasts). P-values can be
computed in two ways: based on a nonparametric Wilcoxon signed-rank test
(internally using wilcox.test() with paired = TRUE) or based on a
permutation test. The permutation test is based on the difference in mean
scores between two models. The default null hypothesis is that the mean score
difference is zero (see permutation_test()).
Adjusted p-values are computed by calling p.adjust() on the raw p-values.
The code for the pairwise comparisons is inspired by an implementation by
Johannes Bracher.
The implementation of the permutation test follows the function
permutationTest from the surveillance package by Michael Höhle,
Andrea Riebler and Michaela Paul.
Usage
get_pairwise_comparisons(
scores,
compare = "model",
by = NULL,
metric = intersect(c("wis", "crps", "brier_score"), names(scores)),
baseline = NULL,
...
)
Arguments
scores |
An object of class |
compare |
Character vector with a single colum name that defines the elements for the pairwise comparison. For example, if this is set to "model" (the default), then elements of the "model" column will be compared. |
by |
Character vector with column names that define further grouping
levels for the pairwise comparisons. By default this is |
metric |
A string with the name of the metric for which a relative skill shall be computed. By default this is either "crps", "wis" or "brier_score" if any of these are available. |
baseline |
A string with the name of a model. If a baseline is
given, then a scaled relative skill with respect to the baseline will be
returned. By default ( |
... |
Additional arguments for the comparison between two models. See
|
Value
A data.table with the results of pairwise comparisons
containing the mean score ratios (mean_scores_ratio),
unadjusted (pval) and adjusted (adj_pval) p-values, and relative skill
values of each model (..._relative_skill). If a baseline model is given
then the scaled relative skill is reported as well
(..._scaled_relative_skill).
Author(s)
Nikos Bosse nikosbosse@gmail.com
Johannes Bracher, johannes.bracher@kit.edu
Examples
library(magrittr) # pipe operator
scores <- example_quantile %>%
as_forecast_quantile() %>%
score()
pairwise <- get_pairwise_comparisons(scores, by = "target_type")
pairwise2 <- get_pairwise_comparisons(
scores, by = "target_type", baseline = "EuroCOVIDhub-baseline"
)
library(ggplot2)
plot_pairwise_comparisons(pairwise, type = "mean_scores_ratio") +
facet_wrap(~target_type)
Probability integral transformation histogram
Description
Generate a Probability Integral Transformation (PIT) histogram for validated forecast objects.
See the examples for how to plot the result of this function.
Usage
## S3 method for class 'forecast_quantile'
get_pit_histogram(forecast, num_bins = NULL, breaks = NULL, by, ...)
## S3 method for class 'forecast_sample'
get_pit_histogram(
forecast,
num_bins = 10,
breaks = NULL,
by,
integers = c("nonrandom", "random", "ignore"),
n_replicates = NULL,
...
)
get_pit_histogram(forecast, num_bins, breaks, by, ...)
## Default S3 method:
get_pit_histogram(forecast, num_bins, breaks, by, ...)
Arguments
forecast |
A forecast object (a validated data.table with predicted and observed values). |
num_bins |
The number of bins in the PIT histogram. For sample-based
forecasts, the default is 10 bins. For quantile-based forecasts, the
default is one bin for each available quantile.
You can control the number of bins by supplying a number. This is fine for
sample-based pit histograms, but may fail for quantile-based formats. In
this case it is preferred to supply explicit breaks points using the
|
breaks |
Numeric vector with the break points for the bins in the
PIT histogram. This is preferred when creating a PIT histogram based on
quantile-based data. Default is |
by |
Character vector with the columns according to which the
PIT values shall be grouped. If you e.g. have the columns 'model' and
'location' in the input data and want to have a PIT histogram for
every model and location, specify |
... |
Currently unused. You cannot pass additional arguments to scoring
functions via |
integers |
How to handle integer forecasts (count data). This is based on methods described Czado et al. (2007). If "nonrandom" (default) the function will use the non-randomised PIT method. If "random", will use the randomised PIT method. If "ignore", will treat integer forecasts as if they were continuous. |
n_replicates |
The number of draws for the randomised PIT for discrete
predictions. Will be ignored if forecasts are continuous or |
Value
A data.table with density values for each bin in the PIT histogram.
References
Sebastian Funk, Anton Camacho, Adam J. Kucharski, Rachel Lowe, Rosalind M. Eggo, W. John Edmunds (2019) Assessing the performance of real-time epidemic forecasts: A case study of Ebola in the Western Area region of Sierra Leone, 2014-15, doi:10.1371/journal.pcbi.1006785
See Also
Examples
library("ggplot2")
result <- get_pit_histogram(example_sample_continuous, by = "model")
ggplot(result, aes(x = mid, y = density)) +
geom_col() +
facet_wrap(. ~ model) +
labs(x = "Quantile", y = "Density")
# example with quantile data
result <- get_pit_histogram(example_quantile, by = "model")
ggplot(result, aes(x = mid, y = density)) +
geom_col() +
facet_wrap(. ~ model) +
labs(x = "Quantile", y = "Density")
Get protected columns from data
Description
Helper function to get the names of all columns in a data frame that are protected columns.
Usage
get_protected_columns(data = NULL)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
Value
A character vector with the names of protected columns in the data.
If data is NULL (default) then it returns a list of all columns that are
protected in scoringutils.
Get interval range belonging to a quantile
Description
Every quantile can be thought of either as the lower or the upper bound of a symmetric central prediction interval. This helper function returns the range of the central prediction interval to which the quantile belongs.
Due to numeric instability that sometimes occurred in the past, ranges are rounded to 10 decimal places. This is not a problem for the vast majority of use cases, but it is something to be aware of.
Usage
get_range_from_quantile(quantile_level)
Arguments
quantile_level |
A numeric vector of quantile levels of size N. |
Value
a numeric vector of interval ranges of size N
Get type of a vector or matrix of observed values or predictions
Description
Internal helper function to get the type of a vector (usually of observed or predicted values). The function checks whether the input is a factor, or else whether it is integer (or can be coerced to integer) or whether it's continuous.
Usage
get_type(x)
Arguments
x |
Input the type should be determined for. |
Value
Character vector of length one with either "classification", "integer", or "continuous".
Illustration of required inputs for binary and point forecasts
Description
Illustration of required inputs for binary and point forecasts
Input format

Overview of required input format for binary and point forecasts
Illustration of required inputs for nominal forecasts
Description
Illustration of required inputs for nominal forecasts
Input format
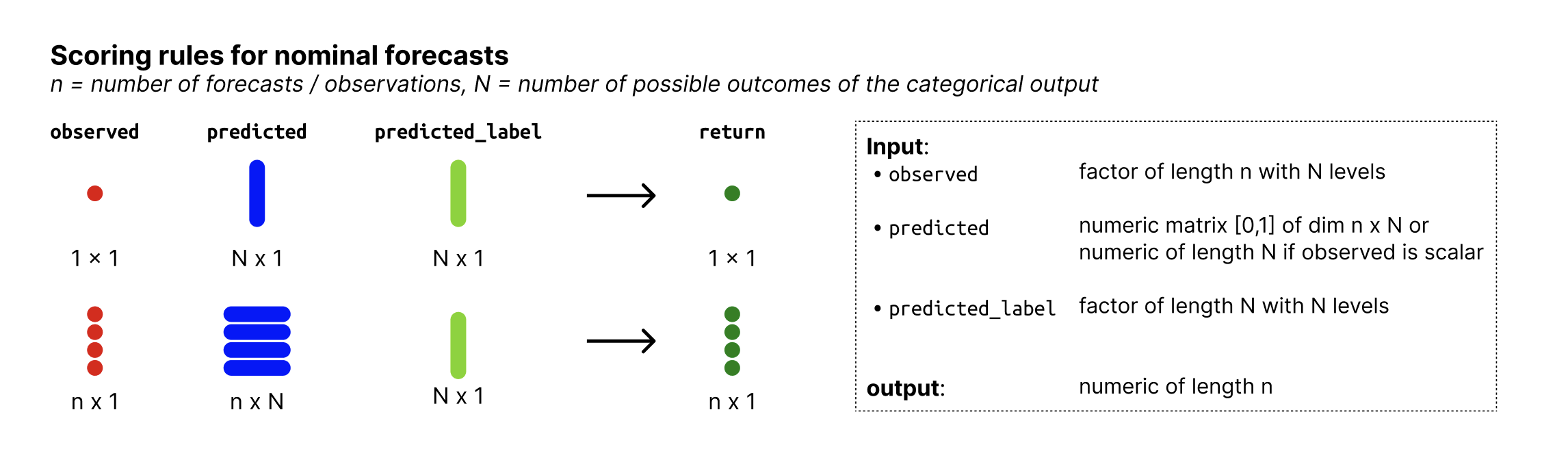
Overview of required input format for nominal forecasts
Illustration of required inputs for ordinal forecasts
Description
Illustration of required inputs for ordinal forecasts
Input format
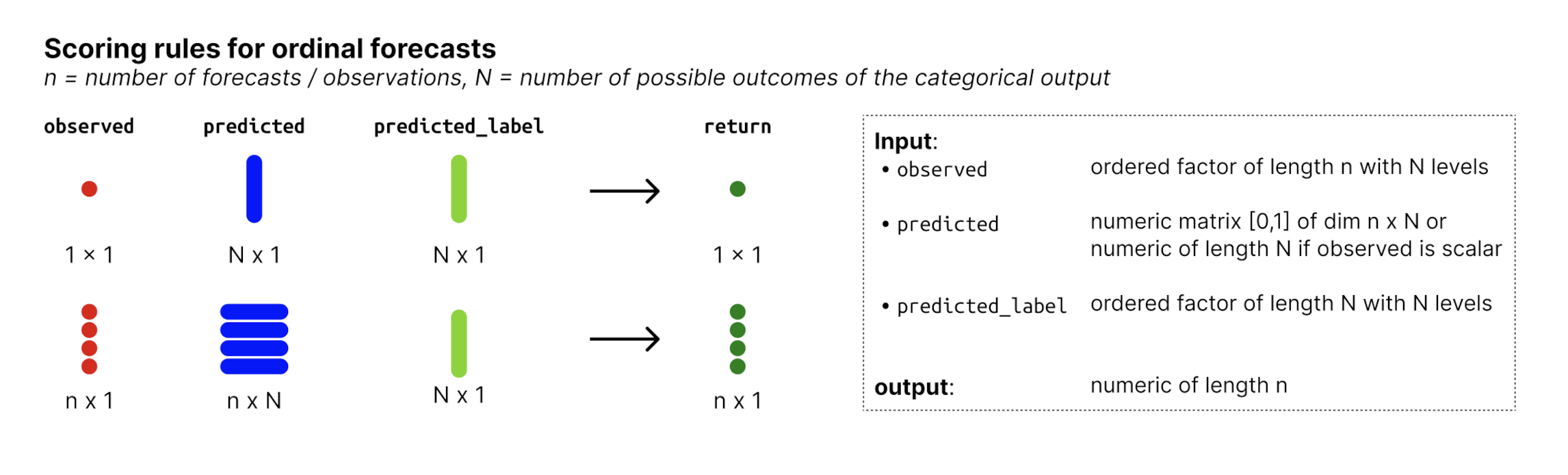
Overview of required input format for ordinal forecasts
Illustration of required inputs for quantile-based forecasts
Description
Illustration of required inputs for quantile-based forecasts
Input format

Overview of required input format for quantile-based forecasts
Illustration of required inputs for sample-based forecasts
Description
Illustration of required inputs for sample-based forecasts
Input format

Overview of required input format for sample-based forecasts
Helper function to interpolate the median prediction if it is not available
Description
Internal function to interpolate the median prediction if it is not available in the given quantile levels. This is done using linear interpolation between the two innermost quantiles.
Usage
interpolate_median(predicted, quantile_level)
Arguments
predicted |
Vector of length N (corresponding to the number of quantiles) that holds predictions. |
quantile_level |
Vector of of size N with the quantile levels for which predictions were made. Note that if this does not contain the median (0.5) then the median is imputed as being the mean of the two innermost quantiles. |
Value
scalar with the imputed median prediction
Input format

Overview of required input format for quantile-based forecasts
Interval coverage (for quantile-based forecasts)
Description
Check whether the observed value is within a given central prediction interval. The prediction interval is defined by a lower and an upper bound formed by a pair of predictive quantiles. For example, a 50% prediction interval is formed by the 0.25 and 0.75 quantiles of the predictive distribution.
Usage
interval_coverage(observed, predicted, quantile_level, interval_range = 50)
Arguments
observed |
Numeric vector of size n with the observed values. |
predicted |
Numeric nxN matrix of predictive
quantiles, n (number of rows) being the number of forecasts (corresponding
to the number of observed values) and N
(number of columns) the number of quantiles per forecast.
If |
quantile_level |
Vector of of size N with the quantile levels for which predictions were made. |
interval_range |
A single number with the range of the prediction interval in percent (e.g. 50 for a 50% prediction interval) for which you want to compute interval coverage. |
Value
A vector of length n with elements either TRUE, if the observed value is within the corresponding prediction interval, and FALSE otherwise.
Input format

Overview of required input format for quantile-based forecasts
Examples
observed <- c(1, -15, 22)
predicted <- rbind(
c(-1, 0, 1, 2, 3),
c(-2, 1, 2, 2, 4),
c(-2, 0, 3, 3, 4)
)
quantile_level <- c(0.1, 0.25, 0.5, 0.75, 0.9)
interval_coverage(observed, predicted, quantile_level)
Interval score
Description
Proper Scoring Rule to score quantile predictions, following Gneiting and Raftery (2007). Smaller values are better.
The score is computed as
\textrm{score} = (\textrm{upper} - \textrm{lower}) + \frac{2}{\alpha}(\textrm{lower}
- \textrm{observed}) *
\mathbf{1}(\textrm{observed} < \textrm{lower}) +
\frac{2}{\alpha}(\textrm{observed} - \textrm{upper}) *
\mathbf{1}(\textrm{observed} > \textrm{upper})
where \mathbf{1}() is the indicator function and
indicates how much is outside the prediction interval.
\alpha is the decimal value that indicates how much is outside
the prediction interval.
To improve usability, the user is asked to provide an interval range in
percentage terms, i.e. interval_range = 90 (percent) for a 90 percent
prediction interval. Correspondingly, the user would have to provide the
5% and 95% quantiles (the corresponding alpha would then be 0.1).
No specific distribution is assumed, but the interval has to be symmetric
around the median (i.e you can't use the 0.1 quantile
as the lower bound and the 0.7 quantile as the upper bound).
Non-symmetric quantiles can be scored using the function quantile_score().
Usage
interval_score(
observed,
lower,
upper,
interval_range,
weigh = TRUE,
separate_results = FALSE
)
Arguments
observed |
A vector with observed values of size n |
lower |
Vector of size n with the prediction for the lower quantile of the given interval range. |
upper |
Vector of size n with the prediction for the upper quantile of the given interval range. |
interval_range |
Numeric vector (either a single number or a vector of
size n) with the range of the prediction intervals. For example, if you're
forecasting the 0.05 and 0.95 quantile, the interval range would be 90.
The interval range corresponds to |
weigh |
Logical. If |
separate_results |
Logical. If |
Value
Vector with the scoring values, or a list with separate entries if
separate_results is TRUE.
References
Strictly Proper Scoring Rules, Prediction,and Estimation, Tilmann Gneiting and Adrian E. Raftery, 2007, Journal of the American Statistical Association, Volume 102, 2007 - Issue 477
Evaluating epidemic forecasts in an interval format, Johannes Bracher, Evan L. Ray, Tilmann Gneiting and Nicholas G. Reich, https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008618 # nolint
Examples
observed <- rnorm(30, mean = 1:30)
interval_range <- rep(90, 30)
alpha <- (100 - interval_range) / 100
lower <- qnorm(alpha / 2, rnorm(30, mean = 1:30))
upper <- qnorm((1 - alpha / 2), rnorm(30, mean = 11:40))
scoringutils:::interval_score(
observed = observed,
lower = lower,
upper = upper,
interval_range = interval_range
)
# gives a warning, as the interval_range should likely be 50 instead of 0.5
scoringutils:::interval_score(
observed = 4, upper = 8, lower = 2, interval_range = 0.5
)
# example with missing values and separate results
scoringutils:::interval_score(
observed = c(observed, NA),
lower = c(lower, NA),
upper = c(NA, upper),
separate_results = TRUE,
interval_range = 90
)
Test whether an object is a forecast object
Description
Test whether an object is a forecast object.
You can test for a specific forecast_<type> class using the appropriate
is_forecast_<type> function.
Usage
is_forecast_binary(x)
is_forecast_nominal(x)
is_forecast_ordinal(x)
is_forecast_point(x)
is_forecast_quantile(x)
is_forecast_sample(x)
is_forecast(x)
Arguments
x |
An R object. |
Value
is_forecast: TRUE if the object is of class forecast,
FALSE otherwise.
is_forecast_<type>*: TRUE if the object is of class forecast_* in addition
to class forecast, FALSE otherwise.
Examples
forecast_binary <- as_forecast_binary(example_binary)
is_forecast(forecast_binary)
Log transformation with an additive shift
Description
Function that shifts a value by some offset and then applies the natural logarithm to it.
Usage
log_shift(x, offset = 0, base = exp(1))
Arguments
x |
vector of input values to be transformed |
offset |
Number to add to the input value before taking the natural logarithm. |
base |
A positive number: the base with respect to which logarithms are computed. Defaults to e = exp(1). |
Details
The output is computed as log(x + offset)
Value
A numeric vector with transformed values
References
Transformation of forecasts for evaluating predictive performance in an epidemiological context Nikos I. Bosse, Sam Abbott, Anne Cori, Edwin van Leeuwen, Johannes Bracher, Sebastian Funk medRxiv 2023.01.23.23284722 doi:10.1101/2023.01.23.23284722 https://www.medrxiv.org/content/10.1101/2023.01.23.23284722v1 # nolint
Examples
library(magrittr) # pipe operator
log_shift(1:10)
log_shift(0:9, offset = 1)
example_quantile[observed > 0, ] %>%
as_forecast_quantile() %>%
transform_forecasts(fun = log_shift, offset = 1)
Log score for categorical outcomes
Description
Log score for categorical (nominal or ordinal) outcomes
The Log Score is the negative logarithm of the probability assigned to the observed value. It is a proper scoring rule. Small values are better (best is zero, worst is infinity).
Usage
logs_categorical(observed, predicted, predicted_label)
Arguments
observed |
Factor of length n with N levels holding the observed values. |
predicted |
nxN matrix of predictive probabilities, n (number of rows) being the number of observations and N (number of columns) the number of possible outcomes. |
predicted_label |
Factor of length N, denoting the outcome
that the probabilities in |
Value
A numeric vector of size n with log scores
Input format

Overview of required input format for nominal forecasts
See Also
Other log score functions:
logs_sample(),
scoring-functions-binary
Examples
factor_levels <- c("one", "two", "three")
predicted_label <- factor(c("one", "two", "three"), levels = factor_levels)
observed <- factor(c("one", "three", "two"), levels = factor_levels)
predicted <- matrix(
c(0.8, 0.1, 0.1,
0.1, 0.2, 0.7,
0.4, 0.4, 0.2),
nrow = 3,
byrow = TRUE
)
logs_categorical(observed, predicted, predicted_label)
Logarithmic score (sample-based version)
Description
This function is a wrapper around the
logs_sample() function from the
scoringRules package.
The log score is the negative logarithm of the predictive density evaluated at the observed value.
The function should be used to score continuous predictions only. While the Log Score is in theory also applicable to discrete forecasts, the problem lies in the implementation: The function uses a kernel density estimation, which is not well defined with integer-valued Monte Carlo Samples. See the scoringRules package for more details and alternatives, e.g. calculating scores for specific discrete probability distributions.
Usage
logs_sample(observed, predicted, ...)
Arguments
observed |
A vector with observed values of size n |
predicted |
nxN matrix of predictive samples, n (number of rows) being
the number of data points and N (number of columns) the number of Monte
Carlo samples. Alternatively, |
... |
Additional arguments passed to logs_sample() from the scoringRules package. |
Value
Vector with scores.
Input format

Overview of required input format for sample-based forecasts
References
Alexander Jordan, Fabian Krüger, Sebastian Lerch, Evaluating Probabilistic Forecasts with scoringRules, https://www.jstatsoft.org/article/view/v090i12
See Also
Other log score functions:
logs_categorical(),
scoring-functions-binary
Examples
observed <- rpois(30, lambda = 1:30)
predicted <- replicate(200, rpois(n = 30, lambda = 1:30))
logs_sample(observed, predicted)
Determine dispersion of a probabilistic forecast
Description
Sharpness is the ability of the model to generate predictions within a narrow range and dispersion is the lack thereof. It is a data-independent measure, and is purely a feature of the forecasts themselves.
Dispersion of predictive samples corresponding to one single observed value is measured as the normalised median of the absolute deviation from the median of the predictive samples. For details, see mad() and the explanations given in Funk et al. (2019)
Usage
mad_sample(observed = NULL, predicted, ...)
Arguments
observed |
Place holder, argument will be ignored and exists only for consistency with other scoring functions. The output does not depend on any observed values. |
predicted |
nxN matrix of predictive samples, n (number of rows) being
the number of data points and N (number of columns) the number of Monte
Carlo samples. Alternatively, |
... |
Additional arguments passed to mad(). |
Value
Vector with dispersion values.
Input format

Overview of required input format for sample-based forecasts
References
Funk S, Camacho A, Kucharski AJ, Lowe R, Eggo RM, Edmunds WJ (2019) Assessing the performance of real-time epidemic forecasts: A case study of Ebola in the Western Area region of Sierra Leone, 2014-15. PLoS Comput Biol 15(2): e1006785. doi:10.1371/journal.pcbi.1006785
Examples
predicted <- replicate(200, rpois(n = 30, lambda = 1:30))
mad_sample(predicted = predicted)
Class constructor for forecast objects
Description
Construct a class based on a data.frame or similar. The constructor
coerces the data into a data.table
assigns a class
Usage
new_forecast(data, classname)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
classname |
name of the class to be created |
Value
An object of the class indicated by classname
Construct an object of class scores
Description
This function creates an object of class scores based on a
data.table or similar.
Usage
new_scores(scores, metrics, ...)
Arguments
scores |
A data.table or similar with scores as produced by |
metrics |
A character vector with the names of the scores (i.e. the names of the scoring rules used for scoring). |
... |
Additional arguments to |
Value
An object of class scores
Examples
## Not run:
df <- data.frame(
model = "A",
wis = "0.1"
)
new_scores(df, "wis")
## End(Not run)
Do pairwise comparison for one set of forecasts
Description
This function does the pairwise comparison for one set of forecasts, but
multiple models involved. It gets called from get_pairwise_comparisons().
get_pairwise_comparisons() splits the data into arbitrary subgroups
specified by the user (e.g. if pairwise comparison should be done separately
for different forecast targets) and then the actual pairwise comparison for
that subgroup is managed from pairwise_comparison_one_group(). In order to
actually do the comparison between two models over a subset of common
forecasts it calls compare_forecasts().
Usage
pairwise_comparison_one_group(
scores,
metric,
baseline,
compare = "model",
by,
...
)
Arguments
scores |
An object of class |
metric |
A string with the name of the metric for which a relative skill shall be computed. By default this is either "crps", "wis" or "brier_score" if any of these are available. |
baseline |
A string with the name of a model. If a baseline is
given, then a scaled relative skill with respect to the baseline will be
returned. By default ( |
compare |
Character vector with a single colum name that defines the elements for the pairwise comparison. For example, if this is set to "model" (the default), then elements of the "model" column will be compared. |
by |
Character vector with column names that define further grouping
levels for the pairwise comparisons. By default this is |
... |
Additional arguments for the comparison between two models. See
|
Value
A data.table with the results of pairwise comparisons
containing the mean score ratios (mean_scores_ratio),
unadjusted (pval) and adjusted (adj_pval) p-values, and relative skill
values of each model (..._relative_skill). If a baseline model is given
then the scaled relative skill is reported as well
(..._scaled_relative_skill).
Simple permutation test
Description
The implementation of the permutation test follows the
function
permutationTest from the surveillance package by Michael Höhle,
Andrea Riebler and Michaela Paul.
The function compares two vectors of scores. It computes the mean of each
vector independently and then takes either the difference or the ratio of
the two. This observed difference or ratio is compared against the same
test statistic based on permutations of the original data.
Used in get_pairwise_comparisons().
Usage
permutation_test(
scores1,
scores2,
n_permutation = 999,
one_sided = FALSE,
comparison_mode = c("difference", "ratio")
)
Arguments
scores1 |
Vector of scores to compare against another vector of scores. |
scores2 |
A second vector of scores to compare against the first |
n_permutation |
The number of replications to use for a permutation test. More replications yield more exact results, but require more computation. |
one_sided |
Whether or not to compute a one-sided test. Default is
|
comparison_mode |
How to compute the test statistic for the comparison of the two scores. Should be either "difference" or "ratio". |
Value
p-value of the permutation test
Probability integral transformation for counts
Description
Uses a Probability integral transformation (PIT) (or a randomised PIT for integer forecasts) to assess the calibration of predictive Monte Carlo samples.
Usage
pit_histogram_sample(
observed,
predicted,
quantiles,
integers = c("nonrandom", "random", "ignore"),
n_replicates = NULL
)
Arguments
observed |
A vector with observed values of size n |
predicted |
nxN matrix of predictive samples, n (number of rows) being
the number of data points and N (number of columns) the number of Monte
Carlo samples. Alternatively, |
quantiles |
A vector of quantiles between which to calculate the PIT. |
integers |
How to handle integer forecasts (count data). This is based on methods described Czado et al. (2007). If "nonrandom" (default) the function will use the non-randomised PIT method. If "random", will use the randomised PIT method. If "ignore", will treat integer forecasts as if they were continuous. |
n_replicates |
The number of draws for the randomised PIT for discrete
predictions. Will be ignored if forecasts are continuous or |
Details
Calibration or reliability of forecasts is the ability of a model to correctly identify its own uncertainty in making predictions. In a model with perfect calibration, the observed data at each time point look as if they came from the predictive probability distribution at that time.
Equivalently, one can inspect the probability integral transform of the predictive distribution at time t,
u_t = F_t (x_t)
where x_t is the observed data point at time t \textrm{ in } t_1,
…, t_n, n being the number of forecasts, and F_t is
the (continuous) predictive cumulative probability distribution at time t. If
the true probability distribution of outcomes at time t is G_t then the
forecasts F_t are said to be ideal if F_t = G_t at all times t.
In that case, the probabilities u_t are distributed uniformly.
In the case of discrete nonnegative outcomes such as incidence counts, the PIT is no longer uniform even when forecasts are ideal. In that case two methods are available ase described by Czado et al. (2007).
By default, a nonrandomised PIT is calculated using the conditional cumulative distribution function
F(u) =
\begin{cases}
0 & \text{if } v < P_t(k_t - 1) \\
(v - P_t(k_t - 1)) / (P_t(k_t) - P_t(k_t - 1)) & \text{if } P_t(k_t - 1) \leq v < P_t(k_t) \\
1 & \text{if } v \geq P_t(k_t)
\end{cases}
where k_t is the observed count, P_t(x) is the predictive
cumulative probability of observing incidence k at time t and
P_t (-1) = 0 by definition.
Values of the PIT histogram are then created by averaging over the n
predictions,
\bar{F}(u) = \frac{i = 1}{n} \sum_{i=1}^{n} F^{(i)}(u)
And calculating the value at each bin between quantile q_i and quantile
q_{i + 1} as
\bar{F}(q_i) - \bar{F}(q_{i + 1})
Alternatively, a randomised PIT can be used instead. In this case, the PIT is
u_t = P_t(k_t) + v * (P_t(k_t) - P_t(k_t - 1))
where v is standard uniform and independent of k. The values of
the PIT histogram are then calculated by binning the u_t values as above.
Value
A vector with PIT histogram densities for the bins corresponding to the given quantiles.
References
Claudia Czado, Tilmann Gneiting Leonhard Held (2009) Predictive model assessment for count data. Biometrika, 96(4), 633-648. Sebastian Funk, Anton Camacho, Adam J. Kucharski, Rachel Lowe, Rosalind M. Eggo, W. John Edmunds (2019) Assessing the performance of real-time epidemic forecasts: A case study of Ebola in the Western Area region of Sierra Leone, 2014-15, doi:10.1371/journal.pcbi.1006785
See Also
Examples
## continuous predictions
observed <- rnorm(20, mean = 1:20)
predicted <- replicate(100, rnorm(n = 20, mean = 1:20))
pit <- pit_histogram_sample(observed, predicted, quantiles = seq(0, 1, 0.1))
## integer predictions
observed <- rpois(20, lambda = 1:20)
predicted <- replicate(100, rpois(n = 20, lambda = 1:20))
pit <- pit_histogram_sample(observed, predicted, quantiles = seq(0, 1, 0.1))
## integer predictions, randomised PIT
observed <- rpois(20, lambda = 1:20)
predicted <- replicate(100, rpois(n = 20, lambda = 1:20))
pit <- pit_histogram_sample(
observed, predicted, quantiles = seq(0, 1, 0.1),
integers = "random", n_replicates = 30
)
Plot correlation between metrics
Description
Plots a heatmap of correlations between different metrics.
Usage
plot_correlations(correlations, digits = NULL)
Arguments
correlations |
A data.table of correlations between scores as produced
by |
digits |
A number indicating how many decimal places the correlations
should be rounded to. By default ( |
Value
A ggplot object showing a coloured matrix of correlations between metrics.
A ggplot object with a visualisation of correlations between metrics
Examples
library(magrittr) # pipe operator
scores <- example_quantile %>%
as_forecast_quantile %>%
score()
correlations <- scores %>%
summarise_scores() %>%
get_correlations()
plot_correlations(correlations, digits = 2)
Visualise the number of available forecasts
Description
Visualise Where Forecasts Are Available.
Usage
plot_forecast_counts(
forecast_counts,
x,
y = "model",
x_as_factor = TRUE,
show_counts = TRUE
)
Arguments
forecast_counts |
A data.table (or similar) with a column |
x |
Character vector of length one that denotes the name of the column to appear on the x-axis of the plot. |
y |
Character vector of length one that denotes the name of the column to appear on the y-axis of the plot. Default is "model". |
x_as_factor |
Logical (default is |
show_counts |
Logical (default is |
Value
A ggplot object with a plot of forecast counts
Examples
library(ggplot2)
library(magrittr) # pipe operator
forecast_counts <- example_quantile %>%
as_forecast_quantile %>%
get_forecast_counts(by = c("model", "target_type", "target_end_date"))
plot_forecast_counts(
forecast_counts, x = "target_end_date", show_counts = FALSE
) +
facet_wrap("target_type")
Create a heatmap of a scoring metric
Description
This function can be used to create a heatmap of one metric across different groups, e.g. the interval score obtained by several forecasting models in different locations.
Usage
plot_heatmap(scores, y = "model", x, metric)
Arguments
scores |
A data.frame of scores based on quantile forecasts as
produced by |
y |
The variable from the scores you want to show on the y-Axis. The default for this is "model" |
x |
The variable from the scores you want to show on the x-Axis. This could be something like "horizon", or "location" |
metric |
String, the metric that determines the value and colour shown in the tiles of the heatmap. |
Value
A ggplot object showing a heatmap of the desired metric
Examples
library(magrittr) # pipe operator
scores <- example_quantile %>%
as_forecast_quantile %>%
score()
scores <- summarise_scores(scores, by = c("model", "target_type"))
scores <- summarise_scores(
scores, by = c("model", "target_type"),
fun = signif, digits = 2
)
plot_heatmap(scores, x = "target_type", metric = "bias")
Plot interval coverage
Description
Plot interval coverage values (see get_coverage() for more information).
Usage
plot_interval_coverage(coverage, colour = "model")
Arguments
coverage |
A data frame of coverage values as produced by
|
colour |
According to which variable shall the graphs be coloured? Default is "model". |
Value
ggplot object with a plot of interval coverage
Examples
example <- as_forecast_quantile(example_quantile)
coverage <- get_coverage(example, by = "model")
plot_interval_coverage(coverage)
Plot heatmap of pairwise comparisons
Description
Creates a heatmap of the ratios or pvalues from a pairwise comparison between models.
Usage
plot_pairwise_comparisons(
comparison_result,
type = c("mean_scores_ratio", "pval")
)
Arguments
comparison_result |
A data.frame as produced by
|
type |
Character vector of length one that is either "mean_scores_ratio" or "pval". This denotes whether to visualise the ratio or the p-value of the pairwise comparison. Default is "mean_scores_ratio". |
Value
A ggplot object with a heatmap of mean score ratios from pairwise comparisons.
Examples
library(ggplot2)
library(magrittr) # pipe operator
scores <- example_quantile %>%
as_forecast_quantile %>%
score()
pairwise <- get_pairwise_comparisons(scores, by = "target_type")
plot_pairwise_comparisons(pairwise, type = "mean_scores_ratio") +
facet_wrap(~target_type)
Plot quantile coverage
Description
Plot quantile coverage values (see get_coverage() for more information).
Usage
plot_quantile_coverage(coverage, colour = "model")
Arguments
coverage |
A data frame of coverage values as produced by
|
colour |
String, according to which variable shall the graphs be coloured? Default is "model". |
Value
A ggplot object with a plot of interval coverage
Examples
example <- as_forecast_quantile(example_quantile)
coverage <- get_coverage(example, by = "model")
plot_quantile_coverage(coverage)
Plot contributions to the weighted interval score
Description
Visualise the components of the weighted interval score: penalties for over-prediction, under-prediction and for high dispersion (lack of sharpness).
Usage
plot_wis(scores, x = "model", relative_contributions = FALSE, flip = FALSE)
Arguments
scores |
A data.table of scores based on quantile forecasts as
produced by |
x |
The variable from the scores you want to show on the x-Axis. Usually this will be "model". |
relative_contributions |
Logical. Show relative contributions instead
of absolute contributions? Default is |
flip |
Boolean (default is |
Value
A ggplot object showing a contributions from the three components of the weighted interval score.
A ggplot object with a visualisation of the WIS decomposition
References
Bracher J, Ray E, Gneiting T, Reich, N (2020) Evaluating epidemic forecasts in an interval format. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008618
Examples
library(ggplot2)
library(magrittr) # pipe operator
scores <- example_quantile %>%
as_forecast_quantile %>%
score()
scores <- summarise_scores(scores, by = c("model", "target_type"))
plot_wis(scores,
x = "model",
relative_contributions = TRUE
) +
facet_wrap(~target_type)
plot_wis(scores,
x = "model",
relative_contributions = FALSE
) +
facet_wrap(~target_type, scales = "free_x")
Print information about a forecast object
Description
This function prints information about a forecast object, including "Forecast type", "Score columns", "Forecast unit".
Usage
## S3 method for class 'forecast'
print(x, ...)
Arguments
x |
A forecast object |
... |
Additional arguments for |
Value
Returns x invisibly.
Examples
dat <- as_forecast_quantile(example_quantile)
print(dat)
Quantile score
Description
Proper Scoring Rule to score quantile predictions. Smaller values are better.
The quantile score is closely related to the interval score (see wis()) and
is the quantile equivalent that works with single quantiles instead of
central prediction intervals.
The quantile score, also called pinball loss, for a single quantile
level \tau is defined as
\text{QS}_\tau(F, y) = 2 \cdot \{ \mathbf{1}(y \leq q_\tau) - \tau\} \cdot (q_\tau - y) =
\begin{cases}
2 \cdot (1 - \tau) * q_\tau - y, & \text{if } y \leq q_\tau\\
2 \cdot \tau * |q_\tau - y|, & \text{if } y > q_\tau,
\end{cases}
with q_\tau being the \tau-quantile of the predictive
distribution F, and \mathbf{1}(\cdot) the indicator function.
The weighted interval score for a single prediction interval can be obtained as the average of the quantile scores for the lower and upper quantile of that prediction interval:
\text{WIS}_\alpha(F, y) = \frac{\text{QS}_{\alpha/2}(F, y)
+ \text{QS}_{1 - \alpha/2}(F, y)}{2}.
See the SI of Bracher et al. (2021) for more details.
quantile_score() returns the average quantile score across the quantile
levels provided. For a set of quantile levels that form pairwise central
prediction intervals, the quantile score is equivalent to the interval score.
Usage
quantile_score(observed, predicted, quantile_level, weigh = TRUE)
Arguments
observed |
Numeric vector of size n with the observed values. |
predicted |
Numeric nxN matrix of predictive
quantiles, n (number of rows) being the number of forecasts (corresponding
to the number of observed values) and N
(number of columns) the number of quantiles per forecast.
If |
quantile_level |
Vector of of size N with the quantile levels for which predictions were made. |
weigh |
Logical. If |
Value
Numeric vector of length n with the quantile score. The scores are
averaged across quantile levels if multiple quantile levels are provided
(the result of calling rowMeans() on the matrix of quantile scores that
is computed based on the observed and predicted values).
Input format

Overview of required input format for quantile-based forecasts
References
Strictly Proper Scoring Rules, Prediction,and Estimation, Tilmann Gneiting and Adrian E. Raftery, 2007, Journal of the American Statistical Association, Volume 102, 2007 - Issue 477
Evaluating epidemic forecasts in an interval format, Johannes Bracher, Evan L. Ray, Tilmann Gneiting and Nicholas G. Reich, 2021, https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008618
Examples
observed <- rnorm(10, mean = 1:10)
alpha <- 0.5
lower <- qnorm(alpha / 2, observed)
upper <- qnorm((1 - alpha / 2), observed)
qs_lower <- quantile_score(observed,
predicted = matrix(lower),
quantile_level = alpha / 2
)
qs_upper <- quantile_score(observed,
predicted = matrix(upper),
quantile_level = 1 - alpha / 2
)
interval_score <- (qs_lower + qs_upper) / 2
interval_score2 <- quantile_score(
observed,
predicted = cbind(lower, upper),
quantile_level = c(alpha / 2, 1 - alpha / 2)
)
# this is the same as the following
wis(
observed,
predicted = cbind(lower, upper),
quantile_level = c(alpha / 2, 1 - alpha / 2)
)
Transform from a quantile format to an interval format
Description
Internal helper function to transform from a quantile format to an interval format (which is no longer a supported forecast format, but still used internally. The function mimics an S3 generic, but is not actually an S3 generic, as we want the functions to be internal and not exported.)
Quantile format In a quantile format, a prediction is characterised by one or multiple predicted values and the corresponding quantile levels. For example, a prediction in a quantile format could be represented by the 0.05, 0.25, 0.5, 0.75 and 0.95 quantiles of the predictive distribution.
Interval format In the interval format, two quantiles are assumed to form a prediction interval. Prediction intervals need to be symmetric around the median and are characterised by a lower and an upper bound. The lower bound is defined by the lower quantile and the upper bound is defined by the upper quantile. A 90% prediction interval, for example, covers 90% of the probability mass and is defined by the 5% and 95% quantiles. A forecast could therefore be characterised by one or multiple prediction intervals, e.g. the lower and upper bounds of the 50% and 90% prediction intervals (corresponding to the 0.25 and 0.75 as well as the 0.05 and 0.095 quantiles).
Usage
quantile_to_interval(...)
quantile_to_interval_dataframe(
forecast,
format = "long",
keep_quantile_col = FALSE,
...
)
quantile_to_interval_numeric(observed, predicted, quantile_level, ...)
Arguments
... |
Arguments |
forecast |
A data.table with forecasts in a quantile-based format (see
|
format |
The format of the output. Either "long" or "wide". If "long"
(the default), there will be a column |
keep_quantile_col |
keep the |
observed |
Numeric vector of size n with the observed values. |
predicted |
Numeric nxN matrix of predictive
quantiles, n (number of rows) being the number of forecasts (corresponding
to the number of observed values) and N
(number of columns) the number of quantiles per forecast.
If |
quantile_level |
Vector of of size N with the quantile levels for which predictions were made. |
Value
A data.table with forecasts in an interval format.
quantile_to_interval_dataframe:
a data.table in an interval format (either "long" or "wide"), with or
without a quantile_level column. Rows will not be reordered.
quantile_to_interval.numeric:
a data.table in a wide interval format with columns forecast_id,
observed, lower, upper, and interval_range. The forecast_id column
is a unique identifier for each forecast. Rows will be reordered according to
forecast_id and interval_range.
Ranked Probability Score for ordinal outcomes
Description
The Ranked Probability Score (RPS) measures the difference between the predicted and observed cumulative distribution functions. It is a proper scoring rule that takes the ordering of categories into account. Small values are better (best is zero, worst is N - 1 where N is the number of categories).
Usage
rps_ordinal(observed, predicted, predicted_label)
Arguments
observed |
A factor of length n with N levels holding the observed values. |
predicted |
nxN matrix of predictive probabilities, n (number of rows) being the number of observations and N (number of columns) the number of possible outcomes. |
predicted_label |
A factor of length N, denoting the outcome that the
probabilities in |
Value
A numeric vector of size n with ranked probability scores
Input format

Overview of required input format for nominal forecasts
Examples
factor_levels <- c("one", "two", "three")
predicted_label <- factor(factor_levels, levels = factor_levels, ordered = TRUE)
observed <- factor(c("three", "three", "two"), levels = factor_levels, ordered = TRUE)
predicted <- matrix(
c(0.8, 0.1, 0.1,
0.1, 0.2, 0.7,
0.4, 0.4, 0.2),
nrow = 3,
byrow = TRUE
)
rps_ordinal(observed, predicted, predicted_label)
Run a function safely
Description
This is a wrapper/helper function designed to run a function safely when it is not completely clear what arguments could be passed to the function.
All named arguments in ... that are not accepted by fun are removed.
All unnamed arguments are passed on to the function. In case fun errors,
the error will be converted to a warning and run_safely returns NULL.
run_safely can be useful when constructing functions to be used as
metrics in score().
Usage
run_safely(..., fun, metric_name)
Arguments
... |
Arguments to pass to |
fun |
A function to execute. |
metric_name |
A character string with the name of the metric. Used to
provide a more informative warning message in case |
Value
The result of fun or NULL if fun errors
Examples
f <- function(x) {x}
scoringutils:::run_safely(2, fun = f, metric_name = "f")
scoringutils:::run_safely(2, y = 3, fun = f, metric_name = "f")
scoringutils:::run_safely(fun = f, metric_name = "f")
scoringutils:::run_safely(y = 3, fun = f, metric_name = "f")
Change data from a sample-based format to a long interval range format
Description
Transform data from a format that is based on predictive samples to a format based on interval ranges.
Usage
sample_to_interval_long(
data,
interval_range = c(0, 50, 90),
type = 7,
keep_quantile_col = TRUE
)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
type |
Type argument passed down to the quantile function. For more
information, see |
keep_quantile_col |
keep quantile_level column, default is TRUE |
Value
A data.table in a long interval interval range format
Evaluate forecasts
Description
score() applies a selection of scoring metrics to a forecast
object.
score() is a generic that dispatches to different methods depending on the
class of the input data.
See as_forecast_binary(), as_forecast_quantile() etc. for information on
how to create a forecast object.
See get_forecast_unit() for more information on the concept of a forecast
unit.
For additional help and examples, check out the paper Evaluating Forecasts with scoringutils in R.
Usage
## S3 method for class 'forecast_binary'
score(forecast, metrics = get_metrics(forecast), ...)
## S3 method for class 'forecast_nominal'
score(forecast, metrics = get_metrics(forecast), ...)
## S3 method for class 'forecast_ordinal'
score(forecast, metrics = get_metrics(forecast), ...)
## S3 method for class 'forecast_point'
score(forecast, metrics = get_metrics(forecast), ...)
## S3 method for class 'forecast_quantile'
score(forecast, metrics = get_metrics(forecast), ...)
## S3 method for class 'forecast_sample'
score(forecast, metrics = get_metrics(forecast), ...)
score(forecast, metrics, ...)
Arguments
forecast |
A forecast object (a validated data.table with predicted and observed values). |
metrics |
A named list of scoring functions. Names will be used as
column names in the output. See |
... |
Currently unused. You cannot pass additional arguments to scoring
functions via |
Details
Customising metrics
If you want to pass arguments to a scoring function, you need change the
scoring function itself via e.g. purrr::partial() and pass an updated list
of functions with your custom metric to the metrics argument in score().
For example, to use interval_coverage() with interval_range = 90, you
would define a new function, e.g.
interval_coverage_90 <- purrr::partial(interval_coverage, interval_range = 90)
and pass this new function to metrics in score().
Note that if you want to pass a variable as an argument, you can
unquote it with !! to make sure the value is evaluated only once when the
function is created. Consider the following example:
custom_arg <- "foo" print1 <- purrr::partial(print, x = custom_arg) print2 <- purrr::partial(print, x = !!custom_arg) custom_arg <- "bar" print1() # prints 'bar' print2() # prints 'foo'
Value
An object of class scores. This object is a data.table with
unsummarised scores (one score per forecast) and has an additional attribute
metrics with the names of the metrics used for scoring. See
summarise_scores()) for information on how to summarise
scores.
Author(s)
Nikos Bosse nikosbosse@gmail.com
References
Bosse NI, Gruson H, Cori A, van Leeuwen E, Funk S, Abbott S (2022) Evaluating Forecasts with scoringutils in R. doi:10.48550/arXiv.2205.07090
Examples
library(magrittr) # pipe operator
validated <- as_forecast_quantile(example_quantile)
score(validated) %>%
summarise_scores(by = c("model", "target_type"))
# set forecast unit manually (to avoid issues with scoringutils trying to
# determine the forecast unit automatically)
example_quantile %>%
as_forecast_quantile(
forecast_unit = c(
"location", "target_end_date", "target_type", "horizon", "model"
)
) %>%
score()
# forecast formats with different metrics
## Not run:
score(as_forecast_binary(example_binary))
score(as_forecast_quantile(example_quantile))
score(as_forecast_point(example_point))
score(as_forecast_sample(example_sample_discrete))
score(as_forecast_sample(example_sample_continuous))
## End(Not run)
Metrics for binary outcomes
Description
Brier score
The Brier Score is the mean squared error between the probabilistic prediction and the observed outcome. The Brier score is a proper scoring rule. Small values are better (best is 0, the worst is 1).
\textrm{Brier\_Score} = (\textrm{prediction} - \textrm{outcome})^2,
where \textrm{outcome} \in \{0, 1\}, and
\textrm{prediction} \in [0, 1] represents
the probability that the outcome is equal to 1.
Log score for binary outcomes
The Log Score is the negative logarithm of the probability assigned to the observed value. It is a proper scoring rule. Small values are better (best is zero, worst is infinity).
Usage
brier_score(observed, predicted)
logs_binary(observed, predicted)
Arguments
observed |
A factor of length n with exactly two levels, holding
the observed values.
The highest factor level is assumed to be the reference level. This means
that |
predicted |
A numeric vector of length n, holding probabilities.
Values represent the probability that the corresponding outcome is equal to
the highest level of the factor |
Details
The functions require users to provide observed values as a factor in order
to distinguish its input from the input format required for scoring point
forecasts. Internally, however, factors will be converted to numeric values.
A factor observed = factor(c(0, 1, 1, 0, 1) with two levels (0 and 1)
would internally be coerced to a numeric vector (in this case this would
result in the numeric vector c(1, 2, 2, 1, 1)). After subtracting 1, the
resulting vector (c(0, 1, 1, 0) in this case) is used for internal
calculations. All predictions are assumed represent the probability that the
outcome is equal of the last/highest factor level (in this case that the
outcome is equal to 1).
You could alternatively also provide a vector like
observed = factor(c("a", "b", "b", "a")) (with two levels, a and b),
which would result in exactly the same internal representation. Probabilities
then represent the probability that the outcome is equal to "b".
If you want your predictions to be probabilities that the outcome is "a",
then you could of course make observed a factor with levels swapped, i.e.
observed = factor(c("a", "b", "b", "a"), levels = c("b", "a"))
Value
A numeric vector of size n with the Brier scores
A numeric vector of size n with log scores
Input format

Overview of required input format for binary and point forecasts
See Also
Other log score functions:
logs_categorical(),
logs_sample()
Examples
observed <- factor(sample(c(0, 1), size = 30, replace = TRUE))
predicted <- runif(n = 30, min = 0, max = 1)
brier_score(observed, predicted)
logs_binary(observed, predicted)
Squared error of the mean (sample-based version)
Description
Squared error of the mean calculated as
\textrm{mean}(\textrm{observed} - \textrm{mean prediction})^2
The mean prediction is calculated as the mean of the predictive samples.
Usage
se_mean_sample(observed, predicted)
Arguments
observed |
A vector with observed values of size n |
predicted |
nxN matrix of predictive samples, n (number of rows) being
the number of data points and N (number of columns) the number of Monte
Carlo samples. Alternatively, |
Input format

Overview of required input format for sample-based forecasts
Examples
observed <- rnorm(30, mean = 1:30)
predicted_values <- matrix(rnorm(30, mean = 1:30))
se_mean_sample(observed, predicted_values)
Select metrics from a list of functions
Description
Helper function to return only the scoring rules selected by the user from a list of possible functions.
Usage
select_metrics(metrics, select = NULL, exclude = NULL)
Arguments
metrics |
A list of scoring functions. |
select |
A character vector of scoring rules to select from the list. If
|
exclude |
A character vector of scoring rules to exclude from the list.
If |
Value
A list of scoring functions.
Examples
select_metrics(
metrics = get_metrics(example_binary),
select = "brier_score"
)
select_metrics(
metrics = get_metrics(example_binary),
exclude = "log_score"
)
Set unit of a single forecast manually
Description
Helper function to set the unit of a single forecast (i.e. the
combination of columns that uniquely define a single forecast) manually.
This simple function keeps the columns specified in forecast_unit (plus
additional protected columns, e.g. for observed values, predictions or
quantile levels) and removes duplicate rows. set_forecast_unit() will
mainly be called when constructing a forecast object
via the forecast_unit argument in as_forecast_<type>.
If not done explicitly, scoringutils attempts to determine the unit
of a single forecast automatically by simply assuming that all column names
are relevant to determine the forecast unit. This may lead to unexpected
behaviour, so setting the forecast unit explicitly can help make the code
easier to debug and easier to read.
Usage
set_forecast_unit(data, forecast_unit)
Arguments
data |
A data.frame (or similar) with predicted and observed values. See the details section of for additional information on the required input format. |
forecast_unit |
Character vector with the names of the columns that uniquely identify a single forecast. |
Value
A data.table with only those columns kept that are relevant to scoring or denote the unit of a single forecast as specified by the user.
Examples
library(magrittr) # pipe operator
example_quantile %>%
scoringutils:::set_forecast_unit(
c("location", "target_end_date", "target_type", "horizon", "model")
)
Summarise scores as produced by score()
Description
Summarise scores as produced by score().
summarise_scores relies on a way to identify the names of the scores and
distinguish them from columns that denote the unit of a single forecast.
Internally, this is done via a stored attribute, metrics that stores
the names of the scores. This means, however, that you need to be careful
with renaming scores after they have been produced by score(). If you
do, you also have to manually update the attribute by calling
attr(scores, "metrics") <- new_names.
Usage
summarise_scores(scores, by = "model", fun = mean, ...)
summarize_scores(scores, by = "model", fun = mean, ...)
Arguments
scores |
An object of class |
by |
Character vector with column names to summarise scores by. Default is "model", i.e. scores are summarised by the "model" column. |
fun |
A function used for summarising scores. Default is |
... |
Additional parameters that can be passed to the summary function
provided to |
Value
A data.table with summarised scores. Scores are summarised according
to the names of the columns of the original data specified in by
using the fun passed to summarise_scores().
Examples
library(magrittr) # pipe operator
scores <- example_sample_continuous %>%
as_forecast_sample() %>%
score()
# get scores by model
summarise_scores(scores, by = "model")
# get scores by model and target type
summarise_scores(scores, by = c("model", "target_type"))
# get standard deviation
summarise_scores(scores, by = "model", fun = sd)
# round digits
summarise_scores(scores, by = "model") %>%
summarise_scores(fun = signif, digits = 2)
Test whether column names are NOT present in a data.frame
Description
The function checks whether all column names are NOT present. If none of the columns are present, the function returns TRUE. If one or more columns are present, the function returns FALSE.
Usage
test_columns_not_present(data, columns)
Arguments
data |
A data.frame or similar to be checked |
columns |
A character vector of column names to check |
Value
Returns TRUE if none of the columns are present and FALSE otherwise
Test whether all column names are present in a data.frame
Description
The function checks whether all column names are present. If one or more columns are missing, the function returns FALSE. If all columns are present, the function returns TRUE.
Usage
test_columns_present(data, columns)
Arguments
data |
A data.frame or similar to be checked |
columns |
A character vector of column names to check |
Value
Returns TRUE if all columns are present and FALSE otherwise
Scoringutils ggplot2 theme
Description
A theme for ggplot2 plots used in scoringutils.
Usage
theme_scoringutils()
Value
A ggplot2 theme
Transform forecasts and observed values
Description
Function to transform forecasts and observed values before scoring.
Usage
transform_forecasts(
forecast,
fun = log_shift,
append = TRUE,
label = "log",
...
)
Arguments
forecast |
A forecast object (a validated data.table with predicted and observed values). |
fun |
A function used to transform both observed values and predictions.
The default function is |
append |
Logical, defaults to |
label |
A string for the newly created 'scale' column to denote the
newly transformed values. Only relevant if |
... |
Additional parameters to pass to the function you supplied. For
the default option of |
Details
There are a few reasons, depending on the circumstances, for why this might be desirable (check out the linked reference for more info). In epidemiology, for example, it may be useful to log-transform incidence counts before evaluating forecasts using scores such as the weighted interval score (WIS) or the continuous ranked probability score (CRPS). Log-transforming forecasts and observations changes the interpretation of the score from a measure of absolute distance between forecast and observation to a score that evaluates a forecast of the exponential growth rate. Another motivation can be to apply a variance-stabilising transformation or to standardise incidence counts by population.
Note that if you want to apply a transformation, it is important to transform the forecasts and observations and then apply the score. Applying a transformation after the score risks losing propriety of the proper scoring rule.
Value
A forecast object with either a transformed version of the data, or one with both the untransformed and the transformed data. includes the original data as well as a transformation of the original data. There will be one additional column, ‘scale’, present which will be set to "natural" for the untransformed forecasts.
Author(s)
Nikos Bosse nikosbosse@gmail.com
References
Transformation of forecasts for evaluating predictive performance in an epidemiological context Nikos I. Bosse, Sam Abbott, Anne Cori, Edwin van Leeuwen, Johannes Bracher, Sebastian Funk medRxiv 2023.01.23.23284722 doi:10.1101/2023.01.23.23284722 https://www.medrxiv.org/content/10.1101/2023.01.23.23284722v1
Examples
library(magrittr) # pipe operator
# transform forecasts using the natural logarithm
# negative values need to be handled (here by replacing them with 0)
example_quantile %>%
.[, observed := ifelse(observed < 0, 0, observed)] %>%
as_forecast_quantile() %>%
# Here we use the default function log_shift() which is essentially the same
# as log(), but has an additional arguments (offset) that allows you add an
# offset before applying the logarithm.
transform_forecasts(append = FALSE) %>%
head()
# alternatively, integrating the truncation in the transformation function:
example_quantile %>%
as_forecast_quantile() %>%
transform_forecasts(
fun = function(x) {log_shift(pmax(0, x))}, append = FALSE
) %>%
head()
# specifying an offset for the log transformation removes the
# warning caused by zeros in the data
example_quantile %>%
as_forecast_quantile() %>%
.[, observed := ifelse(observed < 0, 0, observed)] %>%
transform_forecasts(offset = 1, append = FALSE) %>%
head()
# adding square root transformed forecasts to the original ones
example_quantile %>%
.[, observed := ifelse(observed < 0, 0, observed)] %>%
as_forecast_quantile() %>%
transform_forecasts(fun = sqrt, label = "sqrt") %>%
score() %>%
summarise_scores(by = c("model", "scale"))
# adding multiple transformations
example_quantile %>%
as_forecast_quantile() %>%
.[, observed := ifelse(observed < 0, 0, observed)] %>%
transform_forecasts(fun = log_shift, offset = 1) %>%
transform_forecasts(fun = sqrt, label = "sqrt") %>%
head()
Validate metrics
Description
This function validates whether the list of metrics is a list of valid functions.
The function is used in score() to make sure that all metrics are valid
functions.
Usage
validate_metrics(metrics)
Arguments
metrics |
A named list with metrics. Every element should be a scoring function to be applied to the data. |
Value
A named list of metrics, with those filtered out that are not valid functions
Weighted interval score (WIS)
Description
The WIS is a proper scoring rule used to evaluate forecasts in an interval- / quantile-based format. See Bracher et al. (2021). Smaller values are better.
As the name suggest the score assumes that a forecast comes in the form of one or multiple central prediction intervals. A prediction interval is characterised by a lower and an upper bound formed by a pair of predictive quantiles. For example, a 50% central prediction interval is formed by the 0.25 and 0.75 quantiles of the predictive distribution.
Interval score
The interval score (IS) is the sum of three components: overprediction, underprediction and dispersion. For a single prediction interval only one of the components is non-zero. If for a single prediction interval the observed value is below the lower bound, then the interval score is equal to the absolute difference between the lower bound and the observed value ("underprediction"). "Overprediction" is defined analogously. If the observed value falls within the bounds of the prediction interval, then the interval score is equal to the width of the prediction interval, i.e. the difference between the upper and lower bound. For a single interval, we therefore have:
\textrm{IS} = (\textrm{upper} - \textrm{lower}) + \frac{2}{\alpha}(\textrm{lower}
- \textrm{observed}) *
\mathbf{1}(\textrm{observed} < \textrm{lower}) +
\frac{2}{\alpha}(\textrm{observed} - \textrm{upper}) *
\mathbf{1}(\textrm{observed} > \textrm{upper})
where \mathbf{1}() is the indicator function and
indicates how much is outside the prediction interval.
\alpha is the decimal value that indicates how much is outside
the prediction interval. For a 90% prediction interval, for example,
\alpha is equal to 0.1. No specific distribution is assumed,
but the interval formed by the quantiles has to be symmetric around the
median (i.e you can't use the 0.1 quantile as the lower bound and the 0.7
quantile as the upper bound).
Non-symmetric quantiles can be scored using the function quantile_score().
For a set of k = 1, \dots, K prediction intervals and the median
m, we can compute a weighted interval score (WIS) as the sum of the
interval scores for individual intervals:
\text{WIS}_{\alpha_{\{0:K\}}}(F, y) = \frac{1}{K + 1/2}
\times \left(w_0 \times |y - m| + \sum_{k=1}^{K}
\left\{ w_k \times \text{IS}_{\alpha_k}(F, y) \right\}\right)
The individual scores are usually weighted with
w_k = \frac{\alpha_k}{2}. This weight ensures that
for an increasing number of equally spaced quantiles, the WIS
converges to the continuous ranked probability score (CRPS).
Quantile score
In addition to the interval score, there also exists a quantile score (QS)
(see quantile_score()), which is equal to the so-called pinball loss.
The quantile score can be computed for a single quantile (whereas the
interval score requires two quantiles that form an interval). However,
the intuitive decomposition into overprediction, underprediction and
dispersion does not exist for the quantile score.
Two versions of the weighted interval score
There are two ways to conceptualise the weighted interval score across several quantiles / prediction intervals and the median.
In one view, you would treat the WIS as the average of quantile scores (and
the median as 0.5-quantile) (this is the default for wis()). In another
view, you would treat the WIS as the average of several interval scores +
the difference between the observed value and median forecast. The effect of
that is that in contrast to the first view, the median has twice as much
weight (because it is weighted like a prediction interval, rather than like
a single quantile). Both are valid ways to conceptualise the WIS and you
can control the behaviour with the count_median_twice-argument.
WIS components:
WIS components can be computed individually using the functions
overprediction, underprediction, and dispersion.
Usage
wis(
observed,
predicted,
quantile_level,
separate_results = FALSE,
weigh = TRUE,
count_median_twice = FALSE,
na.rm = FALSE
)
dispersion_quantile(observed, predicted, quantile_level, ...)
overprediction_quantile(observed, predicted, quantile_level, ...)
underprediction_quantile(observed, predicted, quantile_level, ...)
Arguments
observed |
Numeric vector of size n with the observed values. |
predicted |
Numeric nxN matrix of predictive
quantiles, n (number of rows) being the number of forecasts (corresponding
to the number of observed values) and N
(number of columns) the number of quantiles per forecast.
If |
quantile_level |
Vector of of size N with the quantile levels for which predictions were made. |
separate_results |
Logical. If |
weigh |
Logical. If |
count_median_twice |
If TRUE, count the median twice in the score. |
na.rm |
If TRUE, ignore NA values when computing the score. |
... |
Additional arguments passed on to |
Value
wis(): a numeric vector with WIS values of size n (one per observation),
or a list with separate entries if separate_results is TRUE.
dispersion_quantile(): a numeric vector with dispersion values (one per
observation).
overprediction_quantile(): a numeric vector with overprediction values
(one per observation).
underprediction_quantile(): a numeric vector with underprediction values
(one per observation)
Input format

Overview of required input format for quantile-based forecasts
References
Evaluating epidemic forecasts in an interval format, Johannes Bracher, Evan L. Ray, Tilmann Gneiting and Nicholas G. Reich, 2021, https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008618
Examples
observed <- c(1, -15, 22)
predicted <- rbind(
c(-1, 0, 1, 2, 3),
c(-2, 1, 2, 2, 4),
c(-2, 0, 3, 3, 4)
)
quantile_level <- c(0.1, 0.25, 0.5, 0.75, 0.9)
wis(observed, predicted, quantile_level)