| Title: | More Miscellaneous Steps for the 'recipes' Package |
| Version: | 0.3.0 |
| Description: | Contains additional miscellaneous steps for the 'recipes' package. These steps are useful, but doesn't have a good home in other 'recipes' packages or its extensions. |
| License: | MIT + file LICENSE |
| URL: | https://github.com/EmilHvitfeldt/extrasteps, https://emilhvitfeldt.github.io/extrasteps/ |
| BugReports: | https://github.com/EmilHvitfeldt/extrasteps/issues |
| Depends: | R (≥ 3.6), recipes (≥ 1.0.7) |
| Imports: | dplyr, generics, magrittr, purrr, rlang, tibble, vctrs |
| Suggests: | almanac, ggplot2, modeldata, testthat (≥ 3.0.0) |
| Config/testthat/edition: | 3 |
| Encoding: | UTF-8 |
| RoxygenNote: | 7.3.2 |
| NeedsCompilation: | no |
| Packaged: | 2025-09-04 22:03:40 UTC; emilhvitfeldt |
| Author: | Emil Hvitfeldt |
| Maintainer: | Emil Hvitfeldt <emilhhvitfeldt@gmail.com> |
| Repository: | CRAN |
| Date/Publication: | 2025-09-05 16:10:02 UTC |
extrasteps: More Miscellaneous Steps for the 'recipes' Package
Description
Contains additional miscellaneous steps for the 'recipes' package. These steps are useful, but doesn't have a good home in other 'recipes' packages or its extensions.
Author(s)
Maintainer: Emil Hvitfeldt emilhhvitfeldt@gmail.com (ORCID)
See Also
Useful links:
Report bugs at https://github.com/EmilHvitfeldt/extrasteps/issues
Objects exported from other packages
Description
These objects are imported from other packages. Follow the links below to see their documentation.
- generics
- magrittr
S3 methods for tracking which additional packages are needed for steps.
Description
Recipe-adjacent packages always list themselves as a required package so that the steps can function properly within parallel processing schemes.
Usage
## S3 method for class 'step_date_after'
required_pkgs(x, ...)
## S3 method for class 'step_date_before'
required_pkgs(x, ...)
## S3 method for class 'step_date_nearest'
required_pkgs(x, ...)
## S3 method for class 'step_difftime'
required_pkgs(x, ...)
## S3 method for class 'step_encoding_binary'
required_pkgs(x, ...)
## S3 method for class 'step_encoding_frequency'
required_pkgs(x, ...)
## S3 method for class 'step_maxabs'
required_pkgs(x, ...)
## S3 method for class 'step_minmax'
required_pkgs(x, ...)
## S3 method for class 'step_robust'
required_pkgs(x, ...)
## S3 method for class 'step_time_event'
required_pkgs(x, ...)
## S3 method for class 'step_unit_normalize'
required_pkgs(x, ...)
Arguments
x |
A recipe step |
Value
A character vector
Time after Recurrent Date Time Event
Description
step_date_after() creates a specification of a recipe step that will
create new columns indicating the time after an recurrent event.
Usage
step_date_after(
recipe,
...,
role = "predictor",
trained = FALSE,
rules = list(),
transform = "identity",
columns = NULL,
keep_original_cols = FALSE,
skip = FALSE,
id = rand_id("date_after")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose variables for this step.
See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
rules |
Named list of |
transform |
A function or character indication a function used oon the resulting variables. See details for allowed names and their functions. |
columns |
A character string of variables that will be
used as inputs. This field is a placeholder and will be
populated once |
keep_original_cols |
A logical to keep the original variables in the
output. Defaults to |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Details
The transform argument can be function that takes a numeric vector and
returns a numeric vector of the same length. It can also be a character
vector, below is the supported vector names. Some functions come with offset
to avoid Inf.
"identity" function(x) x "inverse" function(x) 1 / (x + 0.5) "exp" function(x) exp(x) "log" function(x) log(x + 0.5)
The effect of transform is illustrated below.
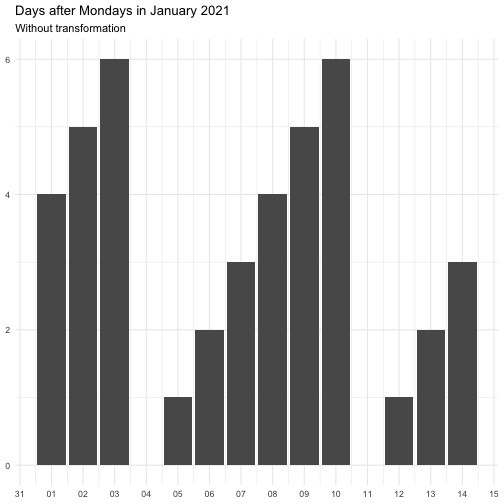
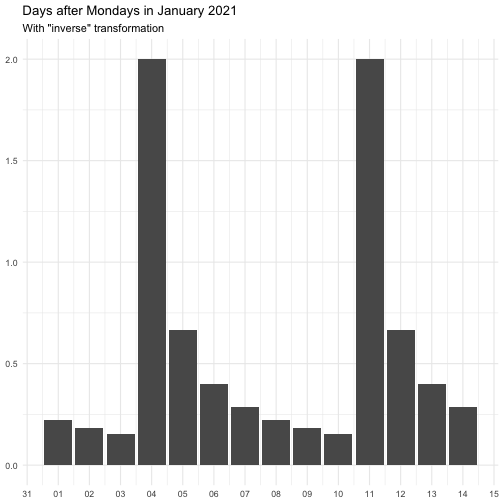
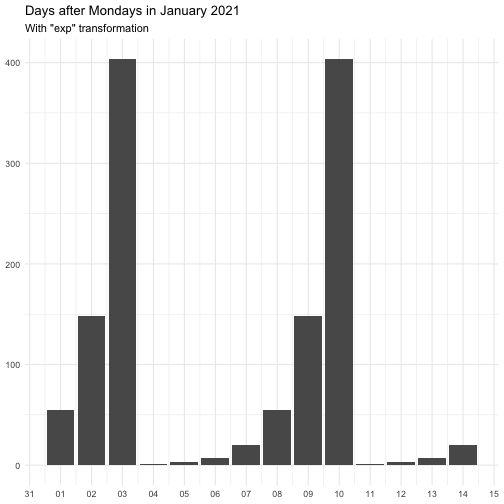
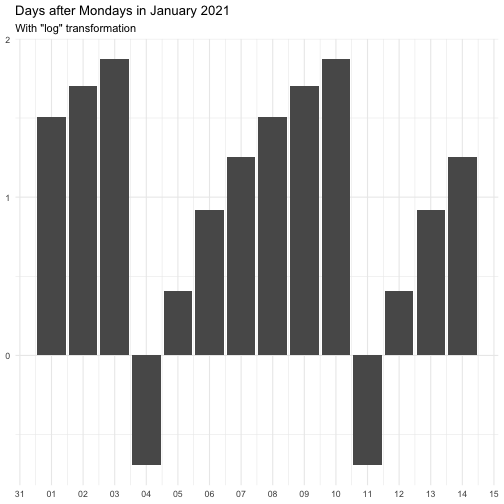
The naming of the resulting variables will be on the form
{variable name}_after_{name of rule}
Value
An updated version of recipe with the new check added to the
sequence of any existing operations.
Examples
library(recipes)
library(extrasteps)
library(almanac)
library(modeldata)
data(Chicago)
on_easter <- yearly() %>% recur_on_easter()
on_weekend <- weekly() %>% recur_on_weekends()
rules <- list(easter = on_easter, weekend = on_weekend)
rec_spec <- recipe(ridership ~ date, data = Chicago) %>%
step_date_after(date, rules = rules)
rec_spec_preped <- prep(rec_spec)
bake(rec_spec_preped, new_data = NULL)
Time before Recurrent Date Time Event
Description
step_date_before() creates a specification of a recipe step that will
create new columns indicating the time before an recurrent event.
Usage
step_date_before(
recipe,
...,
role = "predictor",
trained = FALSE,
rules = list(),
transform = "identity",
columns = NULL,
keep_original_cols = FALSE,
skip = FALSE,
id = rand_id("date_before")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose variables for this step.
See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
rules |
Named list of |
transform |
A function or character indication a function used oon the resulting variables. See details for allowed names and their functions. |
columns |
A character string of variables that will be
used as inputs. This field is a placeholder and will be
populated once |
keep_original_cols |
A logical to keep the original variables in the
output. Defaults to |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Details
The transform argument can be function that takes a numeric vector and
returns a numeric vector of the same length. It can also be a character
vector, below is the supported vector names. Some functions come with offset
to avoid Inf.
"identity" function(x) x "inverse" function(x) 1 / (x + 0.5) "exp" function(x) exp(x) "log" function(x) log(x + 0.5)
The effect of transform is illustrated below.
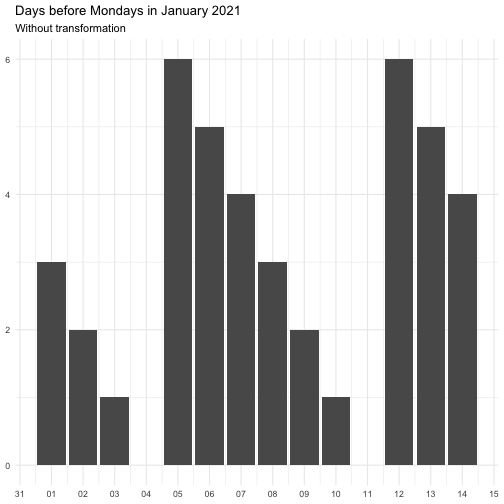
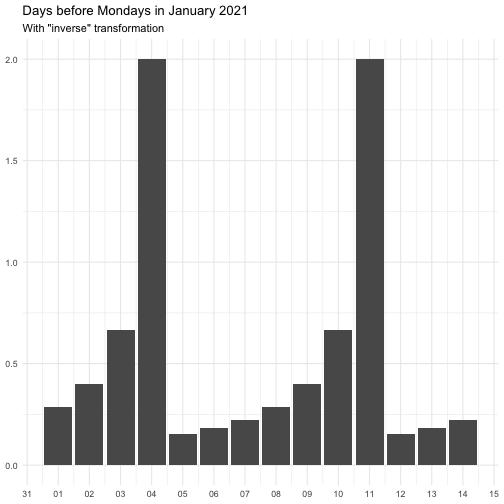
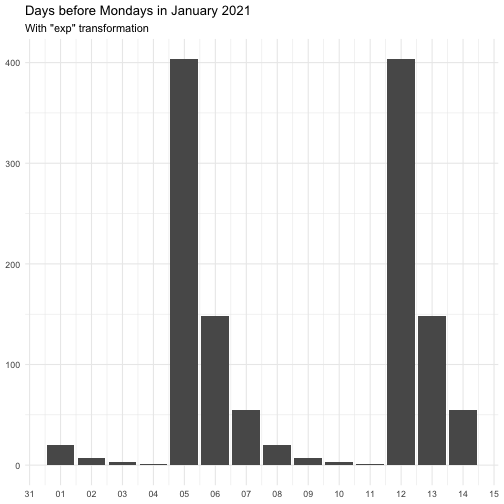
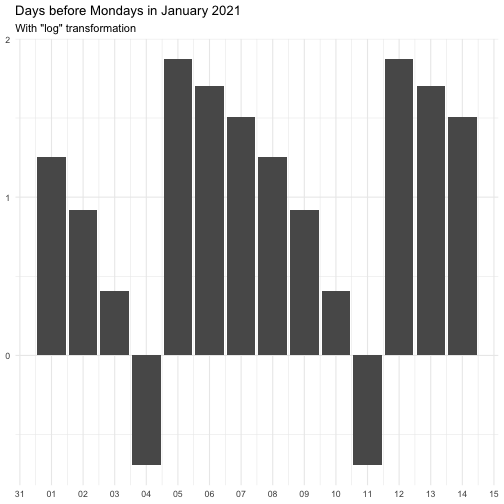
The naming of the resulting variables will be on the form
{variable name}_before_{name of rule}
Value
An updated version of recipe with the new check added to the
sequence of any existing operations.
Examples
library(recipes)
library(extrasteps)
library(almanac)
library(modeldata)
data(Chicago)
on_easter <- yearly() %>% recur_on_easter()
on_weekend <- weekly() %>% recur_on_weekends()
rules <- list(easter = on_easter, weekend = on_weekend)
rec_spec <- recipe(ridership ~ date, data = Chicago) %>%
step_date_before(date, rules = rules)
rec_spec_preped <- prep(rec_spec)
bake(rec_spec_preped, new_data = NULL)
Time to Nearest Recurrent Date Time Event
Description
step_date_nearest() creates a specification of a recipe step that will
create new columns indicating the time to nearest recurrent event.
Usage
step_date_nearest(
recipe,
...,
role = "predictor",
trained = FALSE,
rules = list(),
transform = "identity",
columns = NULL,
keep_original_cols = FALSE,
skip = FALSE,
id = rand_id("date_nearest")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose variables for this step.
See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
rules |
Named list of |
transform |
A function or character indication a function used oon the resulting variables. See details for allowed names and their functions. |
columns |
A character string of variables that will be
used as inputs. This field is a placeholder and will be
populated once |
keep_original_cols |
A logical to keep the original variables in the
output. Defaults to |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Details
The transform argument can be function that takes a numeric vector and
returns a numeric vector of the same length. It can also be a character
vector, below is the supported vector names. Some functions come with offset
to avoid Inf.
"identity" function(x) x "inverse" function(x) 1 / (x + 0.5) "exp" function(x) exp(x) "log" function(x) log(x + 0.5)
The effect of transform is illustrated below.
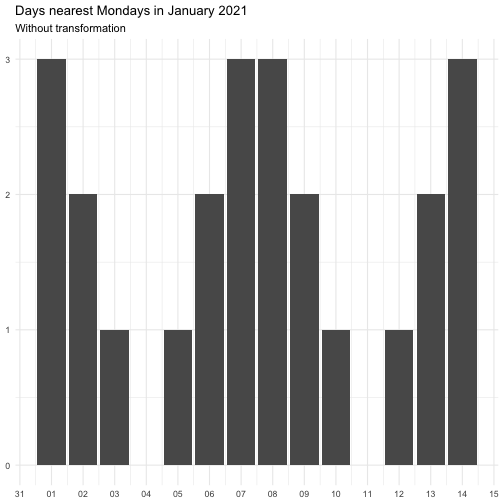
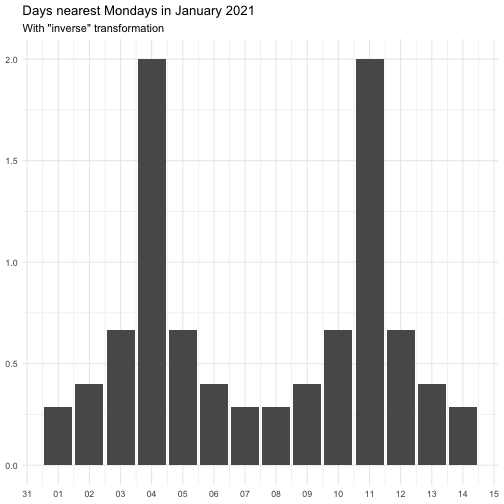
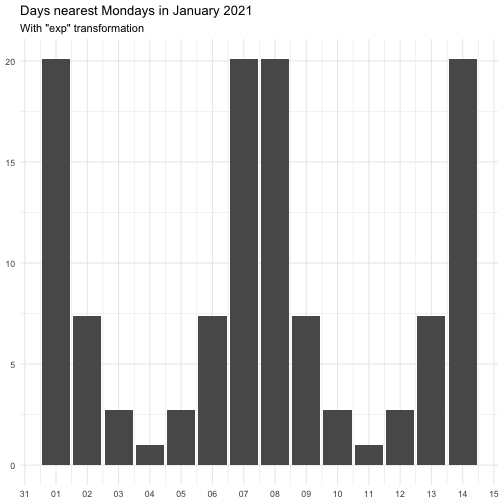
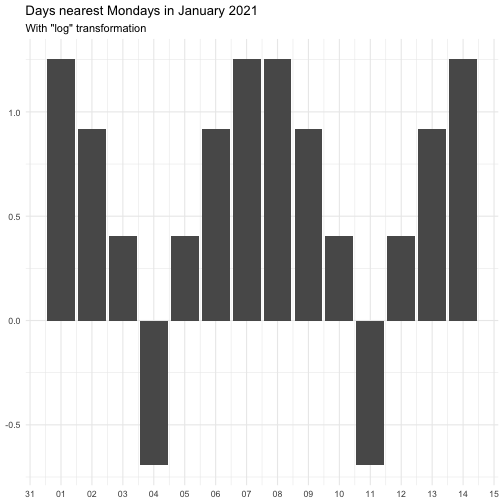
The naming of the resulting variables will be on the form
{variable name}_nearest_{name of rule}
Value
An updated version of recipe with the new check added to the
sequence of any existing operations.
Examples
library(recipes)
library(extrasteps)
library(almanac)
library(modeldata)
data(Chicago)
on_easter <- yearly() %>% recur_on_easter()
on_weekend <- weekly() %>% recur_on_weekends()
rules <- list(easter = on_easter, weekend = on_weekend)
rec_spec <- recipe(ridership ~ date, data = Chicago) %>%
step_date_nearest(date, rules = rules)
rec_spec_preped <- prep(rec_spec)
bake(rec_spec_preped, new_data = NULL)
difftimearithmic Transformation
Description
step_difftime() creates a specification of a recipe step that will
calculate difftimes of the data.
Usage
step_difftime(
recipe,
...,
role = NA,
trained = FALSE,
time = NULL,
tz = NULL,
unit = "auto",
columns = NULL,
skip = FALSE,
id = rand_id("difftime")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose which
variables are affected by the step. See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
time |
date-time or date objects. Used for reference. Must match the type of variable. |
tz |
an optional time zone specification to be used for the conversion, mainly for "POSIXlt" objects. |
unit |
character string. Units in which the results are desired. Must be one of "auto", "secs", "mins", "hours","days", and "weeks" Defaults to "auto". |
columns |
A character string of variable names that will
be populated (eventually) by the |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Value
An updated version of recipe with the new step
added to the sequence of existing steps (if any). For the
tidy method, a tibble with columns terms (the
columns that will be affected) and base.
Examples
library(recipes)
example_date <- data.frame(
dates = seq(as.Date("2010/1/1"), as.Date("2016/1/1"), by = "quarter")
)
example_datetime <- data.frame(
datetimes = seq(ISOdate(1993,1,1), ISOdate(1993,1,2), by = "hour")
)
rec <- recipe(~ dates, data = example_date) %>%
step_difftime(dates, time = as.Date("2010/1/1"))
difftime_obj <- prep(rec)
bake(difftime_obj, new_data = NULL)
recipe(~ dates, data = example_date) %>%
step_difftime(dates, time = as.Date("2010/1/1"), unit = "weeks") %>%
prep() %>%
bake(new_data = NULL)
recipe(~ datetimes, data = example_datetime) %>%
step_difftime(datetimes, time = ISOdate(1993,1,1), unit = "secs") %>%
prep() %>%
bake(new_data = NULL)
Perform binary encoding of factor variables
Description
step_encoding_binary() creates a specification of a recipe step that will
perform binary encoding of factor variables.
Usage
step_encoding_binary(
recipe,
...,
role = NA,
trained = FALSE,
res = NULL,
columns = NULL,
keep_original_cols = FALSE,
skip = FALSE,
id = rand_id("encoding_binary")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose which variables are
affected by the step. See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
res |
A list containing levels of training variables is stored
here once this preprocessing step has be trained by |
columns |
A character string of variable names that will be populated
(eventually) by the |
keep_original_cols |
A logical to keep the original variables in the
output. Defaults to |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Value
An updated version of recipe with the new step added to the
sequence of existing steps (if any). For the tidy method, a tibble with
columns terms (the columns that will be affected) and base.
Examples
library(recipes)
library(modeldata)
data(ames)
rec <- recipe(~ Land_Contour + Neighborhood, data = ames) %>%
step_encoding_binary(all_nominal_predictors()) %>%
prep()
rec %>%
bake(new_data = NULL)
tidy(rec, 1)
Perform frequency encoding
Description
step_encoding_frequency() creates a specification of a recipe step that
will perform frequency encoding.
Usage
step_encoding_frequency(
recipe,
...,
role = NA,
trained = FALSE,
res = NULL,
columns = NULL,
skip = FALSE,
id = rand_id("encoding_frequency")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose which variables are
affected by the step. See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
res |
A list frequencies of the levels of the training variables is
stored here once this preprocessing step has be trained by |
columns |
A character string of variable names that will be populated
(eventually) by the |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Value
An updated version of recipe with the new step added to the
sequence of existing steps (if any). For the tidy method, a tibble with
columns terms (the columns that will be affected) and base.
Examples
library(recipes)
library(modeldata)
data(ames)
rec <- recipe(~ Land_Contour + Neighborhood, data = ames) %>%
step_encoding_frequency(all_nominal_predictors()) %>%
prep()
rec %>%
bake(new_data = NULL)
tidy(rec, 1)
Perform Max Abs Scaling
Description
step_maxabs() creates a specification of a recipe step that will perform
Max Abs scaling.
Usage
step_maxabs(
recipe,
...,
role = NA,
trained = FALSE,
res = NULL,
columns = NULL,
skip = FALSE,
id = rand_id("maxabs")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose which variables are
affected by the step. See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
res |
A list containing absolute max of training variables is stored
here once this preprocessing step has be trained by |
columns |
A character string of variable names that will be populated
(eventually) by the |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Value
An updated version of recipe with the new step added to the
sequence of existing steps (if any). For the tidy method, a tibble with
columns terms (the columns that will be affected) and base.
Examples
library(recipes)
rec <- recipe(~., data = mtcars) %>%
step_maxabs(all_predictors()) %>%
prep()
rec %>%
bake(new_data = NULL)
tidy(rec, 1)
Perform Min Max Scaling
Description
step_minmax() creates a specification of a recipe step that will perform
Min Max scaling.
Usage
step_minmax(
recipe,
...,
role = NA,
trained = FALSE,
res = NULL,
columns = NULL,
skip = FALSE,
id = rand_id("minmax")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose which variables are
affected by the step. See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
res |
A list containing min and max of training variables is stored here
once this preprocessing step has be trained by |
columns |
A character string of variable names that will be populated
(eventually) by the |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Value
An updated version of recipe with the new step added to the
sequence of existing steps (if any). For the tidy method, a tibble with
columns terms (the columns that will be affected) and base.
Examples
library(recipes)
rec <- recipe(~., data = mtcars) %>%
step_minmax(all_predictors()) %>%
prep()
rec %>%
bake(new_data = NULL)
tidy(rec, 1)
Perform Robust Scaling
Description
step_robust() creates a specification of a recipe step that will perform
Robust scaling.
Usage
step_robust(
recipe,
...,
role = NA,
trained = FALSE,
range = c(0.25, 0.75),
res = NULL,
columns = NULL,
skip = FALSE,
id = rand_id("robust")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose which variables are
affected by the step. See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
range |
A numeric vector with 2 values denoting the lower and upper
quantile that is used for scaling. Defaults to |
res |
A list containing the 3 quantiles of training variables is stored
here once this preprocessing step has be trained by |
columns |
A character string of variable names that will be populated
(eventually) by the |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Details
The scaling performed by this step is done using the following transformation
x_new = (x - Q2(x)) / (Q3(x) - Q1(x))
where Q2(x) is the median, Q3(x) is the upper quantile (defaults to 0.75)
and Q1(x) is the lower quantile (defaults to 0.25). The upper and lower
quantiles can be changed with the range argument.
Value
An updated version of recipe with the new step added to the
sequence of existing steps (if any). For the tidy method, a tibble with
columns terms (the columns that will be affected) and base.
Examples
library(recipes)
rec <- recipe(~., data = mtcars) %>%
step_robust(all_predictors()) %>%
prep()
rec %>%
bake(new_data = NULL)
tidy(rec, 1)
rec <- recipe(~., data = mtcars) %>%
step_robust(all_predictors(), range = c(0.1, 0.9)) %>%
prep()
rec %>%
bake(new_data = NULL)
tidy(rec, 1)
Indicate Recurrent Date Time Event
Description
step_time_event() creates a specification of a recipe step that will
create new columns indicating if the date fall on recurrent event.
Usage
step_time_event(
recipe,
...,
role = "predictor",
trained = FALSE,
rules = list(),
columns = NULL,
keep_original_cols = FALSE,
skip = FALSE,
id = rand_id("time_event")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose variables for this step.
See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
rules |
Named list of |
columns |
A character string of variables that will be used as inputs.
This field is a placeholder and will be populated once
|
keep_original_cols |
A logical to keep the original variables in the
output. Defaults to |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Details
Unlike some other steps step_time_event does not remove the
original date variables by default. Set keep_original_cols to FALSE to
remove them.
Value
An updated version of recipe with the new check added to the
sequence of any existing operations.
Examples
library(recipes)
library(extrasteps)
library(almanac)
library(modeldata)
data(Chicago)
on_easter <- yearly() %>% recur_on_easter()
on_weekend <- weekly() %>% recur_on_weekends()
rules <- list(easter = on_easter, weekend = on_weekend)
rec_spec <- recipe(ridership ~ date, data = Chicago) %>%
step_time_event(date, rules = rules)
rec_spec_preped <- prep(rec_spec)
bake(rec_spec_preped, new_data = NULL)
Perform Unit Normalization
Description
step_unit_normalize() creates a specification of a recipe step that will
perform unit normalization by scaling individual samples to have unit norm.
Usage
step_unit_normalize(
recipe,
...,
role = NA,
trained = FALSE,
norm = c("l2", "l1", "max"),
columns = NULL,
skip = FALSE,
id = rand_id("unit_normalize")
)
Arguments
recipe |
A recipe object. The step will be added to the sequence of operations for this recipe. |
... |
One or more selector functions to choose which variables are
affected by the step. See |
role |
Not used by this step since no new variables are created. |
trained |
A logical to indicate if the quantities for preprocessing have been estimated. |
norm |
Character denoting which type of normalization to perform. Must
be one of |
columns |
A character string of variable names that will be populated
(eventually) by the |
skip |
A logical. Should the step be skipped when the recipe is baked by
|
id |
A character string that is unique to this step to identify it. |
Value
An updated version of recipe with the new step added to the
sequence of existing steps (if any). For the tidy method, a tibble with
columns terms (the columns that will be affected) and base.
Examples
library(recipes)
rec <- recipe(~., data = mtcars) %>%
step_unit_normalize(all_predictors()) %>%
prep()
rec %>%
bake(new_data = NULL)
tidy(rec, 1)