| Title: | Multidimensional Cluster Generation Using Support Lines |
| Version: | 1.0.4 |
| Description: | An implementation of the clugen algorithm for generating multidimensional clusters with arbitrary distributions. Each cluster is supported by a line segment, the position, orientation and length of which guide where the respective points are placed. This package is described in Fachada & de Andrade (2023) <doi:10.1016/j.knosys.2023.110836>. |
| Depends: | R (≥ 3.6.0) |
| Imports: | mathjaxr |
| Suggests: | crul, devtools, ggplot2, knitr, lintr, patchwork, prettydoc, rgl, rmarkdown, roxygen2, testthat (≥ 3.0.0) |
| RdMacros: | mathjaxr |
| License: | MIT + file LICENSE |
| URL: | https://clugen.github.io/clugenr/, https://github.com/clugen/clugenr |
| BugReports: | https://github.com/clugen/clugenr/issues |
| Encoding: | UTF-8 |
| RoxygenNote: | 7.3.2 |
| Config/testthat/edition: | 3 |
| VignetteBuilder: | knitr |
| NeedsCompilation: | no |
| Packaged: | 2025-07-07 18:42:55 UTC; nuno |
| Author: | Nuno Fachada |
| Maintainer: | Nuno Fachada <faken@fakenmc.com> |
| Repository: | CRAN |
| Date/Publication: | 2025-07-07 19:00:06 UTC |
clugenr: Multidimensional Cluster Generation Using Support Lines
Description
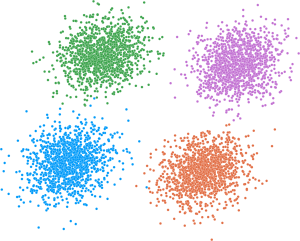
An implementation of the clugen algorithm for generating multidimensional clusters with arbitrary distributions. Each cluster is supported by a line segment, the position, orientation and length of which guide where the respective points are placed. This package is described in Fachada & de Andrade (2023) doi:10.1016/j.knosys.2023.110836.
Author(s)
Maintainer: Nuno Fachada faken@fakenmc.com (ORCID) [copyright holder]
See Also
Useful links:
Report bugs at https://github.com/clugen/clugenr/issues
Angle between two \(n\)-dimensional vectors.
Description
Typically, the angle between two vectors v1 and v2 can be obtained with:
acos((v1 %*% v2) / (norm(v1, "2") * norm(v2, "2")))
However, this approach is numerically unstable. The version provided here is numerically stable and based on the Angle Between Vectors Julia package by Jeffrey Sarnoff (MIT license), implementing an algorithm provided by Prof. W. Kahan in these notes (see page 15).
Usage
angle_btw(v1, v2)
Arguments
v1 |
First vector. |
v2 |
Second vector. |
Value
Angle between v1 and v2 in radians.
Examples
angle_btw(c(1.0, 1.0, 1.0, 1.0), c(1.0, 0.0, 0.0, 0.0)) * 180 / pi
Get angles between average cluster direction and cluster-supporting lines
Description
Determine the angles between the average cluster direction and the
cluster-supporting lines. These angles are obtained from a wrapped normal
distribution (\(\mu=0\), \(\sigma=\) angle_disp ) with
support in the interval \(\left[-\pi/2,\pi/2\right]\).
Note this is different from the standard wrapped normal distribution, the
support of which is given by the interval
\(\left[-\pi,\pi\right]\).
Usage
angle_deltas(num_clusters, angle_disp)
Arguments
num_clusters |
Number of clusters. |
angle_disp |
Angle dispersion, in radians. |
Value
Angles between the average cluster direction and the cluster-supporting lines, given in radians in the interval \(\left[-\pi/2,\pi/2\right]\)
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
set.seed(123)
arad <- angle_deltas(4, pi / 8) # Angle dispersion of 22.5 degrees
arad # What angles deltas did we get?
arad * 180 / pi # Show angle deltas in degrees
Determine cluster centers using the uniform distribution
Description
Determine cluster centers using the uniform distribution, taking into account
the number of clusters (num_clusters) and the average cluster separation
(clu_sep).
More specifically, let \(c=\)num_clusters,
\(\mathbf{s}=\)clu_sep, \(\mathbf{o}=\)clu_offset,
\(n=\)length(clu_sep) (i.e., number of dimensions). Cluster centers
are obtained according to the following equation:
where \(\mathbf{C}\) is the \(c \times n\) matrix of cluster centers, \(\mathbf{U}\) is an \(c \times n\) matrix of random values drawn from the uniform distribution between -0.5 and 0.5, and \(\mathbf{1}\) is an \(c \times 1\) vector with all entries equal to 1.
Usage
clucenters(num_clusters, clu_sep, clu_offset)
Arguments
num_clusters |
Number of clusters. |
clu_sep |
Average cluster separation (\(n \times 1\) vector). |
clu_offset |
Cluster offsets (\(n \times 1\) vector). |
Value
A \(c \times n\) matrix containing the cluster centers.
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
set.seed(321)
clucenters(3, c(30, 10), c(-50,50))
Generate multidimensional clusters
Description
This is the main function of clugenr, and possibly the only function most users will need.
Usage
clugen(
num_dims,
num_clusters,
num_points,
direction,
angle_disp,
cluster_sep,
llength,
llength_disp,
lateral_disp,
allow_empty = FALSE,
cluster_offset = NA,
proj_dist_fn = "norm",
point_dist_fn = "n-1",
clusizes_fn = clusizes,
clucenters_fn = clucenters,
llengths_fn = llengths,
angle_deltas_fn = angle_deltas,
seed = NA
)
Arguments
num_dims |
Number of dimensions. |
num_clusters |
Number of clusters to generate. |
num_points |
Total number of points to generate. |
direction |
Average direction of the cluster-supporting lines. Can be
a vector of length |
angle_disp |
Angle dispersion of cluster-supporting lines (radians). |
cluster_sep |
Average cluster separation in each dimension (vector of
length |
llength |
Average length of cluster-supporting lines. |
llength_disp |
Length dispersion of cluster-supporting lines. |
lateral_disp |
Cluster lateral dispersion, i.e., dispersion of points from their projection on the cluster-supporting line. |
allow_empty |
Allow empty clusters? |
cluster_offset |
Offset to add to all cluster centers (vector of length
|
proj_dist_fn |
Distribution of point projections along cluster-supporting lines, with three possible values:
|
point_dist_fn |
Controls how the final points are created from their projections on the cluster-supporting lines, with three possible values:
|
clusizes_fn |
Distribution of cluster sizes. By default, cluster sizes
are determined by the clusizes function, which uses the normal distribution
(\(\mu=\) |
clucenters_fn |
Distribution of cluster centers. By default, cluster
centers are determined by the clucenters function, which uses the uniform
distribution, and takes into account the |
llengths_fn |
Distribution of line lengths. By default, the lengths of
cluster-supporting lines are determined by the llengths function, which
uses the folded normal distribution (\(\mu=\) |
angle_deltas_fn |
Distribution of line angle differences with respect to
|
seed |
An integer used to initialize the PRNG, allowing for reproducible
results. If specified, |
Details
If a custom function was given in the clusizes_fn parameter, it is
possible that num_points may have a different value than what was
specified in the num_points parameter.
The terms "average" and "dispersion" refer to measures of central tendency and statistical dispersion, respectively. Their exact meaning depends on the optional arguments.
Value
A named list with the following elements:
-
points: Anum_pointsxnum_dimsmatrix with the generated points for all clusters. -
clusters: Anum_pointsfactor vector indicating which cluster each point inpointsbelongs to. -
projections: Anum_pointsxnum_dimsmatrix with the point projections on the cluster-supporting lines. -
sizes: Anum_clustersx 1 vector with the number of points in each cluster. -
centers: Anum_clustersxnum_dimsmatrix with the coordinates of the cluster centers. -
directions: Anum_clustersxnum_dimsmatrix with the final direction of each cluster-supporting line. -
angles: Anum_clustersx 1 vector with the angles between the cluster-supporting lines and the main direction. -
lengths: Anum_clustersx 1 vector with the lengths of the cluster-supporting lines.
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
# 2D example
x <- clugen(2, 5, 1000, c(1, 3), 0.5, c(10, 10), 8, 1.5, 2)
graphics::plot(x$points, col = x$clusters, xlab = "x", ylab = "y", asp = 1)
# 3D example
x <- clugen(3, 5, 1000, c(2, 3, 4), 0.5, c(15, 13, 14), 7, 1, 2)
Merges the fields (specified in fields) of two or more data sets
Description
Merges the fields (specified in fields) of two or more data sets (passed as
lists). The fields to be merged need to have the same number of columns. The
corresponding merged field will contain the rows of the fields to be merged,
and will have a common "supertype".
Usage
clumerge(..., fields = c("points", "clusters"), clusters_field = "clusters")
Arguments
... |
One or more cluster data sets (in the form of lists) whose
|
fields |
Fields to be merged, which must exist in the data sets given in
|
clusters_field |
Field containing the integer cluster labels. If specified, cluster assignments in individual datasets will be updated in the merged dataset so that clusters are considered separate. |
Details
The clusters_field parameter specifies a field containing integers that
identify the cluster to which the respective points belongs to. If
clusters_field is specified (by default it's specified as "clusters"),
cluster assignments in individual datasets will be updated in the merged
dataset so that clusters are considered separate. This parameter can be set
to NA, in which case no field will be considered as a special cluster
assignments field.
This function can be used to merge data sets generated with the clugen
function, by default merging the points and clusters fields in those data
sets. It also works with arbitrary data by specifying alternative fields in
the fields parameter. It can be used, for example, to merge third-party
data with clugen-generated data.
Value
A list whose fields consist of the merged fields in the original data sets.
Examples
a <- clugen(2, 5, 100, c(1, 3), 0.5, c(10, 10), 8, 1.5, 2)
b <- clugen(2, 3, 250, c(-1, 3), 0.5, c(13, 14), 7, 1, 2)
ab <- clumerge(a, b)
Create points from their projections on a cluster-supporting line
Description
Each point is placed around its projection using the normal distribution
(\(\mu=0\), \(\sigma=\) lat_disp ).
Usage
clupoints_n(projs, lat_disp, line_len, clu_dir, clu_ctr)
Arguments
projs |
Point projections on the cluster-supporting line (\(p \times n\) matrix). |
lat_disp |
Standard deviation for the normal distribution, i.e., cluster lateral dispersion. |
line_len |
Length of cluster-supporting line (ignored). |
clu_dir |
Direction of the cluster-supporting line. |
clu_ctr |
Center position of the cluster-supporting line (ignored). |
Details
This function's main intended use is by the main clugen function,
generating the final points when the point_dist_fn parameter is set to
"n".
Value
Generated points (\(p \times n\) matrix).
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
set.seed(123)
ctr <- c(0, 0)
dir <- c(1, 0)
pdist <- c(-0.5, -0.2, 0.1, 0.3)
proj <- points_on_line(ctr, dir, pdist)
clupoints_n(proj, 0.01, NA, dir, NA)
Create points from their projections on a cluster-supporting line
Description
Each point is placed on a hyperplane orthogonal to that line and centered at
the point's projection, using the normal distribution (\(\mu=0\),
\(\sigma=\) lat_disp ).
Usage
clupoints_n_1(projs, lat_disp, line_len, clu_dir, clu_ctr)
Arguments
projs |
Point projections on the cluster-supporting line (\(p \times n\) matrix). |
lat_disp |
Standard deviation for the normal distribution, i.e., cluster lateral dispersion. |
line_len |
Length of cluster-supporting line (ignored). |
clu_dir |
Direction of the cluster-supporting line. |
clu_ctr |
Center position of the cluster-supporting line (ignored). |
Details
This function's main intended use is by the main clugen function,
generating the final points when the point_dist_fn parameter is set to
"n-1".
Value
Generated points (\(p \times n\) matrix).
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
set.seed(123)
ctr <- c(0, 0)
dir <- c(1, 0)
pdist <- c(-0.5, -0.2, 0.1, 0.3)
proj <- points_on_line(ctr, dir, pdist)
clupoints_n_1(proj, 0.1, NA, dir, NA)
Create points from their projections on a cluster-supporting line
Description
Generate points from their \(n\)-dimensional projections on a
cluster-supporting line, placing each point on a hyperplane orthogonal to
that line and centered at the point's projection. The function specified in
dist_fn is used to perform the actual placement.
Usage
clupoints_n_1_template(projs, lat_disp, clu_dir, dist_fn)
Arguments
projs |
Point projections on the cluster-supporting line (\(p \times n\) matrix). |
lat_disp |
Dispersion of points from their projection. |
clu_dir |
Direction of the cluster-supporting line (unit vector). |
dist_fn |
Function to place points on a second line, orthogonal to the first. |
Details
This function is used internally by clupoints_n_1 and may be useful for
constructing user-defined final point placement strategies for the
point_dist_fn parameter of the main clugen function.
Value
Generated points (\(p \times n\) matrix).
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
set.seed(123)
ctr <- c(0, 0)
dir <- c(1, 0)
pdist <- c(-0.5, -0.2, 0.1, 0.3)
proj <- points_on_line(ctr, dir, pdist)
clupoints_n_1_template(proj, 0, dir, function(p, l) stats::runif(p))
Determine cluster sizes, i.e., the number of points in each cluster
Description
Cluster sizes are determined using the normal distribution
(\(\mu=\) num_points \(/\) num_clusters,
\(\sigma=\mu/3\)), and then assuring that the final cluster sizes
add up to num_points via the fix_num_points function.
Usage
clusizes(num_clusters, num_points, allow_empty)
Arguments
num_clusters |
Number of clusters. |
num_points |
Total number of points. |
allow_empty |
Allow empty clusters? |
Value
Number of points in each cluster (vector of length num_clusters).
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
set.seed(123)
sizes <- clusizes(4, 1000, TRUE)
sizes
sum(sizes)
Certify that, given enough points, no clusters are left empty
Description
Certifies that, given enough points, no clusters are left empty. This is done
by removing a point from the largest cluster and adding it to an empty
cluster while there are empty clusters. If the total number of points is
smaller than the number of clusters (or if the allow_empty parameter is set
to TRUE), this function does nothing.
Usage
fix_empty(clu_num_points, allow_empty = FALSE)
Arguments
clu_num_points |
Number of points in each cluster (vector of size \(c\)), where \(c\) is the number of clusters. |
allow_empty |
Allow empty clusters? |
Details
This function is used internally by clusizes and might be useful for custom
cluster sizing implementations given as the clusizes_fn parameter of the
main clugen function.
Value
Number of points in each cluster, after being fixed by this function (vector of size \(c\)).
Examples
clusters <- c(3, 4, 5, 0, 0) # A vector with some empty elements
clusters <- fix_empty(clusters) # Apply this function
clusters # Check that there's no more empty elements
Certify that array values add up to a specific total
Description
Certifies that the values in the clu_num_points array, i.e. the number of
points in each cluster, add up to num_points. If this is not the case, the
clu_num_points array is modified in-place, incrementing the value
corresponding to the smallest cluster while
sum(clu_num_points) < num_points, or decrementing the value corresponding
to the largest cluster while sum(clu_num_points) > num_points.
Usage
fix_num_points(clu_num_points, num_points)
Arguments
clu_num_points |
Number of points in each cluster (vector of size \(c\)), where \(c\) is the number of clusters. |
num_points |
The expected total number of points. |
Details
This function is used internally by clusizes and might be useful for
custom cluster sizing implementations given as the clusizes_fn parameter of
the main clugen function.
Value
Number of points in each cluster, after being fixed by this function.
Examples
clusters <- c(1, 6, 3) # 10 total points
clusters <- fix_num_points(clusters, 12) # But we want 12 total points
clusters # Check that we now have 12 points
Returns the actual dimensions of the input
Description
Gets the dimensions of the input a, returning c(length(a), 1) if
dim(a) == NULL.
Usage
gdim(a)
Determine length of cluster-supporting lines
Description
Line lengths are determined using the folded normal distribution
(\(\mu=\) llength, \(\sigma=\) llength_disp ).
Usage
llengths(num_clusters, llength, llength_disp)
Arguments
num_clusters |
Number of clusters. |
llength |
Average line length. |
llength_disp |
Line length dispersion. |
Value
Lengths of cluster-supporting lines (vector of size num_clusters).
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
set.seed(123)
llengths(4, 20, 3.5)
Determine coordinates of points on a line
Description
Determine coordinates of points on a line with center and direction,
based on the distances from the center given in dist_center.
This works by using the vector formulation of the line equation assuming
direction is a \(n\)-dimensional unit vector. In other words,
considering \(\mathbf{d}=\) as.matrix(direction) (\(n \times
1\) vector), \(\mathbf{c}=\) as.matrix(center) (\(n
\times 1\) vector), and \(\mathbf{w}=\)
as.matrix(dist_center) (\(p \times 1\) vector), the coordinates
of points on the line are given by:
where \(\mathbf{P}\) is the \(p \times n\) matrix of point coordinates on the line, and \(\mathbf{1}\) is a \(p \times 1\) vector with all entries equal to 1.
Usage
points_on_line(center, direction, dist_center)
Arguments
center |
Center of the line (\(n\)-component vector). |
direction |
Line direction (\(n\)-component unit vector). |
dist_center |
Distance of each point to the center of the line (\(n\)-component vector, where \(n\) is the number of points). |
Value
Coordinates of points on the specified line (\(p \times n\) matrix).
Examples
points_on_line(c(5, 5), c(1, 0), seq(-4, 4, length.out=5)) # 2D, 5 points
points_on_line(c(-2, 0, 0, 2), c(0, 0, -1, 0), c(10, -10)) # 4D, 2 points
Get a random unit vector orthogonal to u.
Description
Get a random unit vector orthogonal to u.
Usage
rand_ortho_vector(u)
Arguments
u |
A unit vector. |
Value
A random unit vector orthogonal to u.
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
r <- stats::runif(3) # Get a random 3D vector
r <- r / norm(r, "2") # Normalize it
o <- rand_ortho_vector(r) # Get a random unit vector orthogonal to r
r %*% o # Check that r and o are orthogonal (result should be ~0)
Get a random unit vector with num_dims components.
Description
Get a random unit vector with num_dims components.
Usage
rand_unit_vector(num_dims)
Arguments
num_dims |
Number of components in vector (i.e. vector size). |
Value
A random unit vector with num_dims components.
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
r <- rand_unit_vector(4)
norm(r, "2")
Get a random unit vector at a given angle with another vector.
Description
Get a random unit vector which is at angle radians of vector u.
Note that u is expected to be a unit vector itself.
Usage
rand_vector_at_angle(u, angle)
Arguments
u |
Unit vector with \(n\) components. |
angle |
Angle in radians. |
Value
Random unit vector with \(n\) components which is at angle
radians with vector u.
Note
This function is stochastic. For reproducibility set a PRNG seed with set.seed.
Examples
u <- c(1.0, 0, 0.5, -0.5) # Define a 4D vector
u <- u / norm(u, "2") # Normalize the vector
v <- rand_vector_at_angle(u, pi / 4) # Get a vector at 45 degrees
arad <- acos((u %*% v) / norm(u,"2") * norm(v, "2")) # Get angle in radians
arad * 180 / pi # Convert to degrees, should be close to 45 degrees
Generality of types, by increasing order
Description
Generality of types, by increasing order
Usage
tprom
Format
An object of class list of length 7.