| Type: | Package |
| Title: | Causal Effect for Population Attributable Fractions (PAF) |
| Version: | 1.2.5 |
| Date: | 2022-08-18 |
| Description: | Calculates population attributable fraction causal effects. The 'causalPAF' package contains a suite of functions for causal analysis calculations of population attributable fractions (PAF) given a causal diagram which apply both: Pathway-specific population attributable fractions (PS-PAFs) O’Connell and Ferguson (2022) <doi:10.1093/ije/dyac079> and Sequential population attributable fractions Ferguson, O’Connell, and O’Donnell (2020) <doi:10.1186/s13690-020-00442-x>. Results are presentable in both table and plot format. |
| License: | GPL-2 | GPL-3 [expanded from: GPL (≥ 2)] |
| URL: | https://github.com/MauriceOConnell/causalPAF |
| BugReports: | https://github.com/MauriceOConnell/causalPAF/issues |
| Depends: | R (≥ 2.10) |
| Imports: | checkmate, dagitty, dplyr, forestplot, ggdag, ggplot2, grid, gridExtra, magrittr, MASS, reshape2, rlist, splines, stats, utils |
| Suggests: | spelling |
| Config/testthat/edition: | 3 |
| Encoding: | UTF-8 |
| Language: | en-US |
| LazyData: | true |
| RoxygenNote: | 7.2.1 |
| NeedsCompilation: | no |
| Packaged: | 2022-08-19 23:16:50 UTC; mauriceoconnell |
| Author: | Maurice O'Connell |
| Maintainer: | Maurice O'Connell <maurice.oconnell2@gmail.com> |
| Repository: | CRAN |
| Date/Publication: | 2022-08-19 23:30:02 UTC |
Creates a DAG for input into package Dagitty to identify adjustmentSets given exposure and outcome
Description
Creates a DAG for input into package Dagitty to identify adjustmentSets given exposure and outcome
Usage
addInSplinesTo_in_out(
in_outDAG,
splinesVariables,
in_outDAG_SplinesRemoved,
Splines_outlist_Var,
splinesDefinedIn_in_outDAG,
count,
Subset_adjustmentSet
)
Arguments
in_outDAG |
This defines the causal directed acyclic graph (DAG). A list of length 2. It is defined as a two dimensional list consisting of, firstly, the first list, inlist, i.e. a list of the parents of each variable of interest corresponding to its column name in the data. Splines can be included here if they are to be modelled as splines. Secondly, the second list, outlist, contains a list of a single name of exposure or risk factor or outcome in form of characters i.e. a list of each variable of interest (risk factors, exposures and outcome) corresponding to its column name in the data. Splines should not be input here, only the column names of the variables of interest in the data. The order at which variables are defined must satisfy (i) It is important that variables are defined in the same order in both lists e.g. the first risk factor defined in outlist has its parents listed first in inlist, the second risk factor defined in outlist has its parents listed secondly in inlist and so on. The package assumes this ordering and will not work if this order is violated. (ii) Note it is important also that the order at which the variables are defined is such that all parents of that variable are defined before it. See example in tutorial. |
splinesVariables |
A list of the names of the splines listed in in_outDAG. Note these can only be splines of variables listed in in_out[[2]]. |
in_outDAG_SplinesRemoved |
in_outDAG as defined above but with the splines removed. |
Splines_outlist_Var |
A list defined of same size and order of variables as defined in in_outArg[[2]]. If splines are to be used for variables listed in in_outArg[[2]], then the splines should be defined in Splines_outlist in the same order as variables appear in in_outArg[[2]]. It is necessary to list variables in Splines_outlist the same as in in_outArg[[2]] without splines if no spline is to be applied. It should not be input as an empty list, list(), if no splines. A warning will show if input as an empty list requiring the user to populate Splines_outlist either the same as in_outArg[[2]] (if no splines) or in the same order as in_outArg[[2]] with splines (if splines). See example in tutorial. |
splinesDefinedIn_in_outDAG |
Logical TRUE or FALSE indicating whether the user has defined splines in the causal DAG, in_out, if TRUE. If FALSE and splines are defined in Splines_outlist_Var, then it is necessary for the package to populate the in_out DAG with splines listed in Splines_outlist_Var. |
count |
This variable is calculated within the function make_DAG_AdjustmentSets_in_out.R It is a count variable that should lie somewhere between 0 and length( in_outDAG_SplinesRemoved[[2]] ). If the count is calculated from make_DAG_AdjustmentSets_in_out.R to be equal to length( in_outDAG_SplinesRemoved[[2]] ) then it suggests that all in_outDAG_SplinesRemoved[[1]][[1:length( in_outDAG_SplinesRemoved[[2]] )]] are all valid adjustment sets for each of their outcomes in in_outDAG_SplinesRemoved[[2]] respectively. If count is less than length( in_outDAG_SplinesRemoved[[2]] ) then ( length( in_outDAG_SplinesRemoved[[2]] ) minus count) adjustment sets have been updated in in_outDAG_SplinesRemoved[[1]][[1:length( in_outDAG_SplinesRemoved[[2]] )]] such that they are valid adjustment sets for each of their outcomes in in_outDAG_SplinesRemoved[[2]] respectively. |
Subset_adjustmentSet |
is a list of length length( in_outDAG_SplinesRemoved[[2]] ). It is calculated within the function make_DAG_AdjustmentSets_in_out.R. If all indices of Subset_adjustmentSet[[]] are empty this means that there was no updates to the adjustment sets for each of in_outDAG_SplinesRemoved[[1]][[1:length( in_outDAG_SplinesRemoved[[2]] )]] causal parents of each of in_outDAG_SplinesRemoved[[2]] and this should coincide with count = length( in_outDAG_SplinesRemoved[[2]] ). If count < length( in_outDAG_SplinesRemoved[[2]] ), then ( length( in_outDAG_SplinesRemoved[[2]] ) minus count) adjustment sets have been updated in in_outDAG_SplinesRemoved[[1]][[1:length( in_outDAG_SplinesRemoved[[2]] )]] such that they are valid adjustment sets for each of their outcomes in in_outDAG_SplinesRemoved[[2]] respectively. And the changes to the adjustments sets are stored in each of Subset_adjustmentSet[[1:length( in_outDAG_SplinesRemoved[[2]] )]], where the non-empty index, say i in Subset_adjustmentSet[[i]], corresponds to the variable in in_outDAG_SplinesRemoved[[2]][[i]] that has had its adjustment set in in_outDAG_SplinesRemoved[[1]][[i]] updated. |
Value
Returns in_outDAG with spline variables included
Evaluates Total PAF, Direct PAF, Indirect PAF and Path Specific PAF for a user inputted number of bootstraps and integral simulations
Description
Evaluates Total PAF, Direct PAF, Indirect PAF and Path Specific PAF for a user inputted number of bootstraps and integral simulations
Usage
causalPAFplot(
dataframe,
exposure = "phys",
mediator = c("subhtn", "apob_apoa", "whr"),
response = "case",
response_model_mediators = list(),
response_model_exposure = list(),
in_outArg,
Splines_outlist = list(),
splinesDefinedIn_in_outDAG = list(),
model_listArg = list(),
weights = 1,
NumBootstrap,
NumSimulation,
plot = "bar",
fill = "skyblue",
colour = "orange",
addCustom = FALSE,
custom = ""
)
Arguments
dataframe |
A wide format dataframe containing all the risk factors, confounders, exposures and outcomes within the causal DAG Bayesian network. |
exposure |
The name of the exposure column variable within dataframe in text format e.g. "phys". |
mediator |
The name of the mediator column variables within dataframe in text format. There can be more than one mediator of interest. It can be a vector of mediators names within the dataframe e.g. c("subhtn","apob_apoa","whr"). |
response |
The name of the response column variable within dataframe in text format e.g. "case". The cases should be coded as 1 and the controls as 0. |
response_model_mediators |
A regression model fitted for the response in a causal Bayesian network excluding “children” of the mediators in the causal Bayesian network. See example in tutorial.This model can be listed either as (1) an empty list ( response_model_mediators = list() ) or (2) the user can specify their own customised causal regression model(s) to use. When it is listed as an empty list the 'causalPAF' package will fit the response_model_mediators regression model automatically based on the causal DAG supplied by the user in in_outArg. Alternatively, the user can specify the exact model(s) that the user wishes to use, these model(s) must be in list format (list() where length(response_model_mediators) == length(mediator) ), the same length as the parameter, mediator, with the user customised model for each mediator listed in the same order as in the parameter, mediator, and if there is only one model, it must be listed each time within the list() so that length(response_model_mediators) == length(mediator). |
response_model_exposure |
A regression model fitted for the response in a causal Bayesian network excluding “children” of the exposure in the causal Bayesian network. This regression model will not adjust for mediators (exclude mediators) of the exposure in the regression model so that the total effect of the exposure on the response can be modelled. This model can be listed either as (1) an empty list ( response_model_exposure = list() ) or (2) the user can specify their own customised causal regression model to use. If specified as an empty list, list(), then the causalPAFplot function will define and fit the model automatically based on the causal DAG defined by the in_outArg parameter. Alternatively, the user can specify the exact model that the user wishes to use, this model must be in list format (list() where length(response_model_exposure) == 1 ), of length 1, assuming only one exposure of interest (other exposures can be risk factors) and the model must be defined within a list() since the package assumes a list() format is supplied. See example in tutorial. E.G. If physical exercise ("exer") in the example given in the diagram is the exposure. Then the regression would include all parents of "exer" (i.e. sex, region, educ, age) as well as risk factors at the same level of the causal Bayesian network (i.e. stress, smoke, diet, alcoh). |
in_outArg |
This defines the causal directed acyclic graph (DAG). A list of length 2. It is defined as a two dimensional list consisting of, firstly, the first list, inlist, i.e. a list of the parents of each variable of interest corresponding to its column name in the data. Splines can be included here if they are to be modelled as splines. Secondly, the second list, outlist, contains a list of a single name of exposure or risk factor or outcome in form of characters i.e. a list of each variable of interest (risk factors, exposures and outcome) corresponding to its column name in the data. Splines should not be input here, only the column names of the variables of interest in the data. The order at which variables are defined must satisfy (i) It is important that variables are defined in the same order in both lists e.g. the first risk factor defined in outlist has its parents listed first in inlist, the second risk factor defined in outlist has its parents listed secondly in inlist and so on. The package assumes this ordering and will not work if this order is violated. (ii) Note it is important also that the order at which the variables are defined is such that all parents of that variable are defined before it. See example in tutorial. |
Splines_outlist |
A list defined of same size and order of variables as defined in in_outArg[[2]]. If splines are to be used for variables listed in in_outArg[[2]], then the splines should be defined in Splines_outlist in the same order as variables appear in in_outArg[[2]]. It is necessary to list variables in Splines_outlist the same as in in_outArg[[2]] without splines if no spline is to be applied. It should not be input as an empty list, list(), if no splines. A warning will show if input as an empty list requiring the user to populate Splines_outlist either the same as in_outArg[[2]] (if no splines) or in the same order as in_outArg[[2]] with splines (if splines). See example in tutorial. |
splinesDefinedIn_in_outDAG |
Logical TRUE or FALSE indicating whether the user has defined splines in the causal DAG, in_out, if TRUE. If FALSE and splines are defined in Splines_outlist_Var, then it is necessary for the package to populate the in_out DAG with splines listed in Splines_outlist_Var. |
model_listArg |
is a list of models fitted for each of the variables in in_outArg[[2]] (or in_outArg |
weights |
Column of weights for case control matching listed in the same order as the patients in the data e.g. weights = strokedata$weights. |
NumBootstrap |
The number of bootstraps the user wants to use to calculate confidence intervals for the effect. A minimum of 200 bootstrap replications (Efron (2016), Computer Age Statistical Inference, page 162) are recommended to calculate standard errors (for intervals of the form: estimate plus or minus 1.96*(standard error of bootstrap estimate. However increasing the number of bootstraps can result in the package taking a long time to run. So the user may decide to balance speed with accuracy depending on which is of more value in the specific context. |
NumSimulation |
This is the number of simulations requested by the user to estimate integrals. The larger the number of simulations the more accurate the results but the longer the code takes to run. Therefore the user may wish to balance speed with accuracy depending on which is of more value in the specific context of interest. The integrals for continuous variables are estimated using simulation methods. |
plot |
plot can be text inputs "forestplot" or "bar" where:"forestplot" plots a forest plot."bar" plots a bar chart with error bars. |
fill |
The colour for the fill in the bar chart is set here in text format. The default is fill= "skyblue". |
colour |
The colour for the error bar in the bar chart is set here in text format. The default is colour = "orange". |
addCustom |
Logical TRUE or FALSE indicating whether a customised interaction term is to be added to the each regression. The interaction term can include splines. |
custom |
text containing the customised interaction term to be added to each regression. The text should be enclosed in inverted commas. Splines can be included within the interaction terms. See tutorial for examples. |
Value
Prints a forest plot or a bar chart with error bars of the 5 results for each mediator. The 5 results are:(1)Total Population Attributable Fraction (PAF),(2)Direct Effect Population Attributable Fraction (PAF) using alternative definition, (3)Indirect Effect Population Attributable Fraction (PAF) using alternative definition, (4)Path Specific Population Attributable Fraction (PAF), (5)Overall Direct Population Attributable Fraction (PAF)
Examples
# Loads some data (fictional Stroke data from the package 'causalPAF')
# In this example, we use a small data set called 'strokedata_smallSample' consisting of 5,000
# rows of fictional patient data. For more accurate results, a larger data set is available
# called 'strokedata'which contains 16,623 rows of fictional patient data. The methodology
# applied in the 'causalPAF' package is more accurate the larger the dataset. To use the larger
# 'strokedata' dataset, simply call
# stroke_reduced <- strokedata
stroke_reduced <- strokedata_smallSample
# Just shortening the name of a variable, "apob_apoa", to "apb" so the R code
# in document example is not truncated.
stroke_reduced$apb <- stroke_reduced$apob_apoa
# The data should contain a column of weights for case control matching.
# strokedata$weights
# Weigths are not needed for cohort/cross sectional designs.
# The data should have reference levels of all risk factors already set.
# This can be done as follows but has already been applied to the data so is not run here:
# levels(stroke_reduced$htnadmbp) <- c(0, 1)
# stroke_reduced$subhtn <- factor(stroke_reduced$subhtn,levels=c(1, 2))
# levels(stroke_reduced$nevfcur) <- c(1, 2)
# stroke_reduced$global_stress2 <- factor(stroke_reduced$global_stress2,levels=c(1,2))
# levels(stroke_reduced$whrs2tert) <- c(1, 2, 3)
# levels(stroke_reduced$phys) <- c(2, 1)
# levels(stroke_reduced$alcohfreqwk) <- c(1, 2, 3)
# stroke_reduced$dmhba1c2 <- factor(stroke_reduced$dmhba1c2,levels=c(1,2))
# stroke_reduced$cardiacrfcat <- factor(stroke_reduced$cardiacrfcat,levels=c(1,2))
# levels(stroke_reduced$ahei3tert) <- c(3,2,1)
# levels(stroke_reduced$apob_apoatert) <- c(1,2,3)
# The 'causalPAF' package assumes the data is either complete case data or that missing data
# analysis has already been performed.
# Next, define the causal structure or directed acyclic graph (DAG) of the causal Bayesian
# network defined by the data. We list the parents of each exposure or risk factor or outcome
# in a vector as follows:
# Note it is important that the order at which the variables are defined is such that all
# parents of that variable are defined before it. Please refer to the figure of the causal
# Bayesian network (with both direct and indirect effects) defined earlier as an example of this
# order.
in_phys <- c("subeduc","moteduc","fatduc")
in_ahei <- c("subeduc","moteduc","fatduc")
in_nevfcur <- c("subeduc","moteduc","fatduc")
in_alcohfreqwk <- c("subeduc","moteduc","fatduc")
in_global_stress2 <- c("subeduc","moteduc","fatduc")
in_subhtn <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_apob_apoa <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_whr <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
# Note splines can be fitted within the causal structure as shown below especially if splines
# are to be used in the fitted models.
# It is important that splines of parent variables are "typed" or "spelt" consistently
# (including spaces) throughout as 'causalPAF' can fit models automatically provided variables are
# spelt consistently. Also if a parent variable is a spline it should be defined in spline
# format in all occurences of the parent variable.
in_cardiacrfcat <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2",
"ns(apb,knots=quantile(apb,c(.25,.5,.75)),Boundary.knots=quantile(apb,c(.001,.95)))",
"ns(whr,df=5)","subhtn")
in_dmhba1c2 <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2",
"ns(apb,knots=quantile(apb,c(.25,.5,.75)),Boundary.knots=quantile(apb,c(.001,.95)))",
"ns(whr,df=5)","subhtn")
in_case <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2",
"ns(apb,knots=quantile(apb,c(.25,.5,.75)),Boundary.knots=quantile(apb,c(.001,.95)))",
"ns(whr,df=5)","subhtn","cardiacrfcat","dmhba1c2")
# Then we define a two dimensional list consisting of
# 1. inlist i.e. a list of the parents of each variable of interest corresponding to its column
# name in the data. Splines should be included here if they are to be modelled as splines.
# 2. outlist i.e. a list of each variable of interest corresponding to its column name in the
# data. Splines should not be input here, only the column names of the variables of interest in
# the data.
# Again the order is such that each variable is defined after all its parents.
in_out <- list(inlist=list(in_phys,in_ahei,in_nevfcur,in_alcohfreqwk,in_global_stress2,
in_subhtn,in_apob_apoa,in_whr,in_cardiacrfcat,in_dmhba1c2,in_case),
outlist=c("phys","ahei3tert","nevfcur","alcohfreqwk","global_stress2","subhtn",
"apb","whr","cardiacrfcat","dmhba1c2","case"))
# If splines are to be used for variables listed in in_out$outlist, then the splines should be
# defined in the same order as variables appear in in_out$outlist as follows. It is necessary to
# list variables in in_out$outlist without splines if no spline is to be applied.
# It is important that Splines_outlist is defined in the following format
# list(c("splinename1","splinename2","splinename3")) for the package to be applied correctly.
# And Splines_outlist should not be an empty list(). If there are no splines it should be
# defined the same as in_out[[2]] and in the same order as variables defined in_out[[2]].
Splines_outlist = list( c("phys","ahei3tert","nevfcur","alcohfreqwk","global_stress2","subhtn",
"ns(apb,knots=quantile(apb,c(.25,.5,.75)),Boundary.knots=quantile(apb,c(.001,.95)))",
"ns(whr,df=5)","cardiacrfcat","dmhba1c2","case") )
# To fit these models to case control data, one needs to perform weighted maximum likelihood
# estimation to imitate estimation using a random sample from the population. We chose weights
# of 0.0035 (for each case) and 0.9965 (for each control), reflective of a yearly incidence of
# first ischemic stroke of 0.35%, or 3.5 strokes per 1,000 individuals. These weights were
# chosen according to average incidences across country, age, group and gender within
# INTERSTROKE according to the global burden of disease.
w <- rep(1,nrow(stroke_reduced))
w[stroke_reduced$case==0] <- 0.9965
w[stroke_reduced$case==1] <- 0.0035
# It is important to assign stroke_reduced$weights to the updated weights defined in w.
# Otherwise if stroke_reduced$weights <- w is not set, the alternative weights supplied in the
# fictional data will be used. In this case, we want to use weigths as defined in w.
stroke_reduced$weights <- w
#The checkMarkovDAG() function in the 'causalPAF' package should be used before running
# causalPAFplot() to ensure:
#1. The causal Markov condition holds for the causal structure defined in the variable in_out.
#2. The variables in in_out are listed in the order so that no variable is defined before a
# parent or direct cause. Note: if this order does not hold, checkMarkovDAG() will automatically
# reorder the variables in, in_out, provided it is a Markov DAG.
#The causal analysis requires that the causal structure is a Markov DAG. The Causal Markov (CM)
# condition states that, conditional on the set of all its direct causes, a node is independent
# of all variables which are not direct causes or direct effects of that node. In the event that
# the structure of a Bayesian network accurately depicts causality, the two conditions are
# equivalent. However, a network may accurately embody the Markov condition without depicting
# causality, in which case it should not be assumed to embody the causal Markov condition.
# in_out is as defined above and input into this code.
if(checkMarkovDAG(in_out)$IsMarkovDAG & !checkMarkovDAG(in_out)$Reordered){
print("Your in_out DAG is a Markov DAG.")
} else if( checkMarkovDAG(in_out)$IsMarkovDAG & checkMarkovDAG(in_out)$Reordered ) {
in_out <- checkMarkovDAG(in_out)[[2]]
print("Your in_out DAG is a Markov DAG.The checkMarkovDAG function has reordered your
in_out list so that all parent variables come before descendants.")
} else{ print("Your ``in_out'' list is not a Bayesian Markov DAG so the methods in the
'causalPAF' package cannot be applied for non Markov DAGs.")}
# The pointEstimate() function evaluates Point Estimates for Total PAF, Direct PAF, Indirect PAF
# and Path Specific PAF for a user inputted number of integral simulations. There is no bootstrap
# applied in this fucntion.
# Since bootstraps are not applied, the pointEstimate() function will run quicker than the
# alternative causalPAFplot() function which calculates bootstrap estimates which can take
# longer to run.
pointEstimate(dataframe = stroke_reduced,
exposure="phys",
mediator=c("subhtn","apb","whr"),
response="case",
response_model_mediators = list(),
response_model_exposure = list(),
in_outArg = in_out,
Splines_outlist = Splines_outlist,
splinesDefinedIn_in_outDAG = TRUE,
model_listArg = list(),
weights = w,
NumSimulation = 3,
addCustom = TRUE,
custom = "regionnn7*ns(eage,df=5)+esex*ns(eage,df=5)")
# The causalPAFplot() function will perform Pathway Specific Population Attributable Fraction
# (PSPAF) calculations and output results based on an exposure, mediators and response input
# by the user according to the columns names of these variables defined in the dataframe.
# Setting model_listArg, response_model_mediators and response_model_exposure by default to an
# empty list will instruct the 'causalPAF' package to fit these models automatically based on the
# causal DAG supplied in the in _outArg. Alternatively the user can supply their custom fitted,
# model_listpop, response_model_mediators and response_model_exposure which should be consistent
# with the causal structure.
# Note we fit a custom interaction for the outcome (or case or response) regression
# ( custom = "regionnn7*ns(eage,df=5)+esex*ns(eage,df=5)") ). Care should be taken that the
# customised regression should not contain variables that might affect the causal interpretation
# of the regression e.g. in this case we have used baseline confounders (i.e. regionn, eage and
# esex) with interactions and splines. In general, using baseline confounders in custom should
# not affect any causal interpretations whereas using variables far ``downstream'' might block
# causal pathways. The user is required to apply discretion in using ``addCustom'' and
# ``Custom'' in ensuring a causal interpretation remains. If no customisation is required the
# user can input addCustom = FALSE and custom = "" which is the default setting.
# Finally we call the causalPAFplot function for the pathway specific PAF calculations as
# follows:
# For greater accuracy a larger number of bootstraps (e.g. 200) and larger number of simulations
# (e.g. 1000) should be run. However, this will increase the run time greatly.
causalPAFplot(dataframe = stroke_reduced,
exposure="phys",
mediator=c("subhtn","apb","whr"),
response="case",
response_model_mediators = list(),
response_model_exposure = list(),
in_outArg = in_out,
Splines_outlist = Splines_outlist,
splinesDefinedIn_in_outDAG = TRUE,
model_listArg = list(),
weights = w,
NumBootstrap = 2,
NumSimulation = 2,
plot = "bar",
fill= "skyblue",
colour="orange",
addCustom = TRUE,
custom = "regionnn7*ns(eage,df=5)+esex*ns(eage,df=5)")
# The causalPAFplot function below has response_model_mediators, response_model_exposure and
# model_listArg pre fit. This allows the user to apply customised regressions instead of the
# default setting above, where the 'causalPAF' R package fitted these regressions automatically
# based on the causalDAG defined in in_outArg.
# Libraries must be loaded if fitting models outside of the 'causalPAF' R package.
library(MASS)
library(splines)
# Next we fit the, response_model_mediators and response_model_exposure, models outside of the
# 'causalPAF' package as an input into the package.
# It is important that response_vs_mediator is a list and it must be the same length as the
# parameter, mediator, i.e. length( response_vs_mediator ) == length( mediator). In this
# example, mediator=c("subhtn","apb","whr") so length( mediator) is 3, so we create a list
# with three models for "subhtn","apb" and "whr" respectively in that order. Note in this
# example, the model is the same for each mediator, but it must still be input 3 times within
# the list as follows:
response_vs_mediator <- list(
glm("case ~ regionnn7*ns(eage,df=5)+esex*ns(eage,df=5) +subeduc+moteduc+ fatduc+ phys+
ahei3tert+ nevfcur+ alcohfreqwk+ global_stress2+ subhtn +
ns(apb, knots = quantile(apb,c(.25,0.5,0.75)),
Boundary.knots = quantile(apb,c(.001,0.95)))+
ns(whr,df=5)",data = stroke_reduced,family='binomial',w = stroke_reduced$weights ),
# "subhtn" mediator model
glm("case ~ regionnn7*ns(eage,df=5)+esex*ns(eage,df=5) + subeduc+ moteduc+ fatduc+ phys+
ahei3tert+ nevfcur+ alcohfreqwk+ global_stress2+ subhtn +
ns(apb, knots = quantile(apb,c(.25,0.5,0.75)),
Boundary.knots = quantile(apb,c(.001,0.95)))+
ns(whr,df=5)",data = stroke_reduced,family='binomial',w = stroke_reduced$weights ),
# "apob_apoa" mediator model name shoretd to "apb"
glm("case ~ regionnn7*ns(eage,df=5)+esex*ns(eage,df=5) + subeduc+ moteduc+ fatduc+ phys+
ahei3tert+ nevfcur+ alcohfreqwk+ global_stress2+ subhtn +
ns(apb, knots = quantile(apb,c(.25,0.5,0.75)),
Boundary.knots = quantile(apb,c(.001,0.95)))+
ns(whr,df=5)",data = stroke_reduced,family='binomial',w = stroke_reduced$weights ) )
# "whr" mediator model
# Next we fit a customised response_model_exposure model rather than allowing the package fit it
# automatically as shown previously. This must be a list of length 1.
response_vs_phys <- list(glm("case ~ regionnn7*ns(eage,df=5)+esex*ns(eage,df=5)+subeduc+moteduc+
fatduc+ phys+ ahei3tert+ nevfcur+ alcohfreqwk+ global_stress2",data = stroke_reduced,
family='binomial',w= stroke_reduced$weights) )
# model_listArg is a list of models fitted for each of the variables in in_out$outlist based on
# its parents given in in_out$inlist. By default this is set to an empty list. Alternatively the
# user can supply their custom fitted, model_listpop, which should be consistent with the causal
# structure. model_listArg is defined earlier in this tutorial.
# Note it is important that model_listArg is defined as a list and in the same order and length
# as the variables defined in in_outArg[[2]].
model_listArgFit <- list(glm(formula = phys ~ subeduc + regionnn7 * ns(eage, df = 5) +
esex * ns(eage, df = 5) + moteduc + fatduc, family = "binomial", data = stroke_reduced,
weights = weights), # model 1 phys
polr(formula = ahei3tert ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, data = stroke_reduced, weights = weights), # model 2 ahei3tert
glm(formula = nevfcur ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, family = "binomial",data = stroke_reduced, weights = weights),
# model 3 nevfcur
polr(formula = alcohfreqwk ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc, data = stroke_reduced,weights = weights), # model 4 alcohfreqwk
glm(formula = global_stress2 ~ subeduc + regionnn7 * ns(eage,df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, family = "binomial",data = stroke_reduced, weights = weights),
# model 5 global_stress2
glm(formula = subhtn ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2,family = "binomial",
data = stroke_reduced, weights = weights), # model 6 subhtn
lm(formula = apb ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2,
data = stroke_reduced,weights = weights), # model 7 apob_apoa name shorted to "apb"
lm(formula = whr ~ subeduc + regionnn7 * ns(eage, df = 5) + esex *ns(eage, df = 5) + moteduc +
fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2, data = stroke_reduced,
weights = weights), # model 8 whr
glm(formula = cardiacrfcat ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2 +
ns(apb, knots = quantile(apb,c(0.25, 0.5, 0.75)),
Boundary.knots = quantile(apb,c(0.001, 0.95))) + ns(whr, df = 5) + subhtn,
family = "binomial",data = stroke_reduced, weights = weights), # model 9 cardiacrfcat
glm(formula = dmhba1c2 ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2 +
ns(apb, knots = quantile(apb,c(0.25, 0.5, 0.75)),
Boundary.knots = quantile(apb,c(0.001, 0.95))) + ns(whr, df = 5) + subhtn,
family = "binomial",data = stroke_reduced, weights = weights), # model 10 dmhba1c2
glm(formula = case ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) + moteduc +
fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2 +
ns(apb, knots = quantile(apb,c(0.25, 0.5, 0.75)),
Boundary.knots = quantile(apb,c(0.001, 0.95))) + ns(whr, df = 5) + subhtn +
cardiacrfcat +dmhba1c2, family = "binomial", data = stroke_reduced, weights = weights)
# model 11 case
)
# For greater accuracy a larger number of bootstraps (e.g. 200) and larger number of simulations
# (e.g. 1000) should be run. However, this will increase the run time greatly.
causalPAFplot(dataframe = stroke_reduced,
exposure="phys",
mediator=c("subhtn","apb","whr"),
response="case",
response_model_mediators = response_vs_mediator,
response_model_exposure = response_vs_phys,
in_outArg = in_out,
Splines_outlist = Splines_outlist,
splinesDefinedIn_in_outDAG = TRUE,
model_listArg = model_listArgFit,
weights = w,
NumBootstrap = 2,
NumSimulation = 2,
plot = "bar",
fill= "skyblue",
colour ="orange" )
Checks if the causal DAG satisfies the Markov condition
Description
The functions checks if the Markov condition holds for the Directed Acyclic Graph (DAG) defined. Sometimes called the Markov assumption, is an assumption made in Bayesian probability theory, that every node in a Bayesian network is conditionally independent of its nondescendants, given its parents. In other words, it is assumed that a node has no bearing on nodes which do not descend from it. This is equivalent to stating that a node is conditionally independent of the entire network, given its Markov blanket. The related Causal Markov condition states that, conditional on the set of all its direct causes, a node is independent of all variables which are not direct causes or direct effects of that node.
Usage
checkMarkovDAG(in_out)
Arguments
in_out |
A list of length 2. The first list contains a list of character vectors of the parents of the exposure or risk factor or outcome which are either causes or confounders of the exposure or risk factor or outcome. The second list contains a list of a single name of exposure or risk factor or outcome in form of characters. See tutorial examples for examples. |
Value
IsMarkovDAG |
Returns a logical TRUE or FALSE whether it is a Markov DAG provided in_out is input as described in the documentation. |
in_out |
The in_out list supplied in the function is returns the same of the input if IsMarkovDAG is returned TRUE. If IsMarkovDAG is returned FALSE the order of the in_out list is updated such that all parent variables come before ancestors in both i_out[[1]] and in_out[[2]]. This corrects any error where variables from a given Markov Bayesian DAG are input to the package in the incorrect order. |
Reordered |
Reordered is FALSE if in_out is left in the same order as input. Reordered is FALSE if in_out has been reordered so that parents of variables could before descendants. |
Examples
# Loads some data (fictional Stroke data from the package 'causalPAF')
# In this example, we use a small data set called 'strokedata_smallSample' consisting of 5,000
# rows of fictional patient data. For more accurate results, a larger data set is available
# called 'strokedata'which contains 16,623 rows of fictional patient data. The methodology
# applied in the 'causalPAF' package is more accurate the larger the dataset. To use the larger
# 'strokedata' dataset, simply call
# stroke_reduced <- strokedata
stroke_reduced <- strokedata_smallSample
in_phys <- c("subeduc","moteduc","fatduc")
in_ahei <- c("subeduc","moteduc","fatduc")
in_nevfcur <- c("subeduc","moteduc","fatduc")
in_alcohfreqwk <- c("subeduc","moteduc","fatduc")
in_global_stress2 <- c("subeduc","moteduc","fatduc")
in_htnadmbp <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_apob_apoatert <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_whrs2tert <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_cardiacrfcat <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2", "apob_apoatert","whrs2tert","htnadmbp")
in_dmhba1c2 <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2", "apob_apoatert","whrs2tert","htnadmbp")
in_case <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2", "apob_apoatert","whrs2tert","htnadmbp","cardiacrfcat","dmhba1c2")
in_out <- list(inlist=list(in_phys,in_ahei,in_nevfcur,in_alcohfreqwk,in_global_stress2,
in_htnadmbp, in_apob_apoatert,in_whrs2tert,in_cardiacrfcat,
in_dmhba1c2,in_case),
outlist=c("phys","ahei3tert","nevfcur","alcohfreqwk","global_stress2",
"htnadmbp","apob_apoatert", "whrs2tert","cardiacrfcat",
"dmhba1c2","case"))
if(checkMarkovDAG(in_out)$IsMarkovDAG & !checkMarkovDAG(in_out)$Reordered){
print("Your in_out DAG is a Markov DAG.")
} else if( checkMarkovDAG(in_out)$IsMarkovDAG & checkMarkovDAG(in_out)$Reordered ) {
in_out <- checkMarkovDAG(in_out)[[2]]
print("Your in_out DAG is a Markov DAG.The checkMarkovDAG function has reordered your
in_out list so that all parent variables come before descendants.")
} else{ print("Your ``in_out'' list is not a Bayesian Markov DAG so the methods in the
'causalPAF' package cannot be applied for non Markov DAGs.")}
Makes Formula
Description
Given a causal
Usage
do_sim(model, dataframe)
Arguments
model |
A fitted mediator regression model |
dataframe |
Data frame containing data to be analysed. The data frame has cases in rows and variables in columns. |
Value
dataframe |
dataframe |
Examples
## Not run:
# I don't want you to run this
## End(Not run)
Simulates a Fitted Model for a Mediator or Exposure or Risk Factor Allowing for Potential Outcomes in Causal Analysis
Description
A fitted model for a mediator or exposure or risk factor can be simulated given values of the other risk
factors or exposure saved in the data frame current_mat. This allows for potential outcomes to be measured for
causal analysis. For example, for an outcome Y_{A,M} with exposure A and mediators M_{1}, M_{3}, \dots M_{K}
the function can measure potential outcomes such as Y_{A=0,M_{1},M_{2},M_{3}} or Y_{A=0,M_{1},M_{2}=0,M_{3}=0} when there are three mediators.
The model can be either a binary, continuous or an ordered factor response model.
Usage
do_sim_sequentialPAF(colnum, current_mat, model)
Arguments
colnum |
Column number of exposure or risk factor of interest within the data frame. The data frame has cases in rows and variables in columns. |
current_mat |
The data frame containing the data for which the model can be simulated with. For
potential outcomes for example such as |
model |
A fitted causal regression model for either a binary, continuous or an ordered factor response. |
Value
simulation |
simulation |
Examples
## Not run:
# I don't want you to run this
## End(Not run)
Evaluates and Makes Formula for regression of exposure or risk factor or outcome on its parents in a causal Bayesian network directed acyclic graph.
Description
Evaluates and Makes Formula for regression of exposure or risk factor or outcome on its parents in a causal Bayesian network directed acyclic graph. Given a causal Bayesian network, directed acyclic graph (DAG) where arrows representing
causal dependencies between confounders, risk factors,exposure and disease, together with a sensible probability distribution on
the graph that respects these causal dependencies. To consistently estimate causal effects that risk factors may have on each
other and on disease, we need to make a strong no unmeasured confounding assumption: that is common causes of nodes in the graph,
which may be causes of two risk factors or a cause of risk factor and disease, are also included as nodes in the graph.
Causal Bayesian networks have a local Markov property that the conditional probability distribution of any node X_j, given values
for the other variables in the network, only depends on the values x_{pa_{j}} of the parent nodes.
Usage
eval_make_DAG(
data,
regressionExposure,
regressionMediator,
response,
response_model_mediators,
response_model_exposure,
w
)
Arguments
data |
A wide format data containing all the risk factors, confounders, exposures and outcomes within the causal DAG Bayesian network. |
regressionExposure |
Regression of response given exposure based on adjustment set output from function make_DAG.R. |
regressionMediator |
Regression of response given exposure (mediator as exposure) based on canonical adjustment set output from function make_DAG.R. |
response |
The name of the response column variable within dataframe in text format e.g. “case”. The cases should be coded as 1 and the controls as 0. |
response_model_mediators |
A model fitted for the response in a causal Bayesian network excluding “children” of the mediators in the causal Bayesian network. See example in tutorial. |
response_model_exposure |
A model fitted for the response in a causal Bayesian network excluding “children” of the exposure and risk factors in the causal Bayesian network. See example in tutorial. |
w |
Column of weights for case control matching listing in same order as patients in data. |
Value
regressionExposure_listReturn model list regressionExposure_listReturn
regressionMediator_listReturn model list regressionMediator_listReturn
Evaluates and Makes Formula for regression of exposure or risk factor or outcome on its parents in a causal Bayesian network directed acyclic graph.
Description
Evaluates and Makes Formula for regression of exposure or risk factor or outcome on its parents in a causal Bayesian network directed acyclic graph. Given a causal Bayesian network, directed acyclic graph (DAG) where arrows representing
causal dependencies between confounders, risk factors, exposure and disease, together with a sensible probability distribution on
the graph that respects these causal dependencies. To consistently estimate causal effects that risk factors may have on each
other and on disease, we need to make a strong no unmeasured confounding assumption: that is common causes of nodes in the graph,
which may be causes of two risk factors or a cause of risk factor and disease, are also included as nodes in the graph.
Causal Bayesian networks have a local Markov property that the conditional probability distribution of any node X_j, given values
for the other variables in the network, only depends on the values x_{pa_{j}} of the parent nodes.
Usage
eval_make_formula(data, in_out, model_list, w, addCustom = FALSE, custom = "")
Arguments
data |
A wide format data containing all the risk factors, confounders, exposures and outcomes within the causal DAG Bayesian network. |
in_out |
This defines the causal directed acyclic graph (DAG). A list of length |
model_list |
is a list of models fitted for each of the variables in in_outArg[[2]] (or in_outArg |
w |
Column of weights for case control matching listing in same order as patients in data. |
addCustom |
Logical TRUE or FALSE indicating whether a customised interaction term is to be added to the each regression. The interaction term can include splines. |
custom |
text containing the customised interaction term to be added to each regression. The text should be enclosed in inverted commas. Splines can be included within the interaction terms. See tutorial for examples. |
Value
model_listReturn[[1]] model list A
model_listReturn[[2]] model list B
model_listReturn[[3]] model list C
model_listReturn[[4]] model list D
model_listReturn[[5]] model list E
model_listReturn[[6]] model list F
model_listReturn[[7]] model list G
model_listReturn[[8]] model list H
model_listReturn[[9]] model list I
model_listReturn[[10]] model list J
model_listReturn[[11]] model list K
Calculation of Population Attributable Fraction (PAF), with a decomposition of the total PAF into direct and indirect components.
Description
Calculation of Population Attributable Fraction (PAF), with a decomposition of the total PAF into direct
and indirect components. It performs one simulation which can be combined with a bootstrap approach to
perform multiple simulations. If we think of Y_0 as the potential outcome for
an individual if they were never exposed to the risk factor, can be directly interpreted as the relative change in
disease prevalence if an exposure was absent from the population.
Sjolander introduced the ideas of mediation into the literature for PAF, defining a decomposition of the total PAF
into direct and indirect components: and
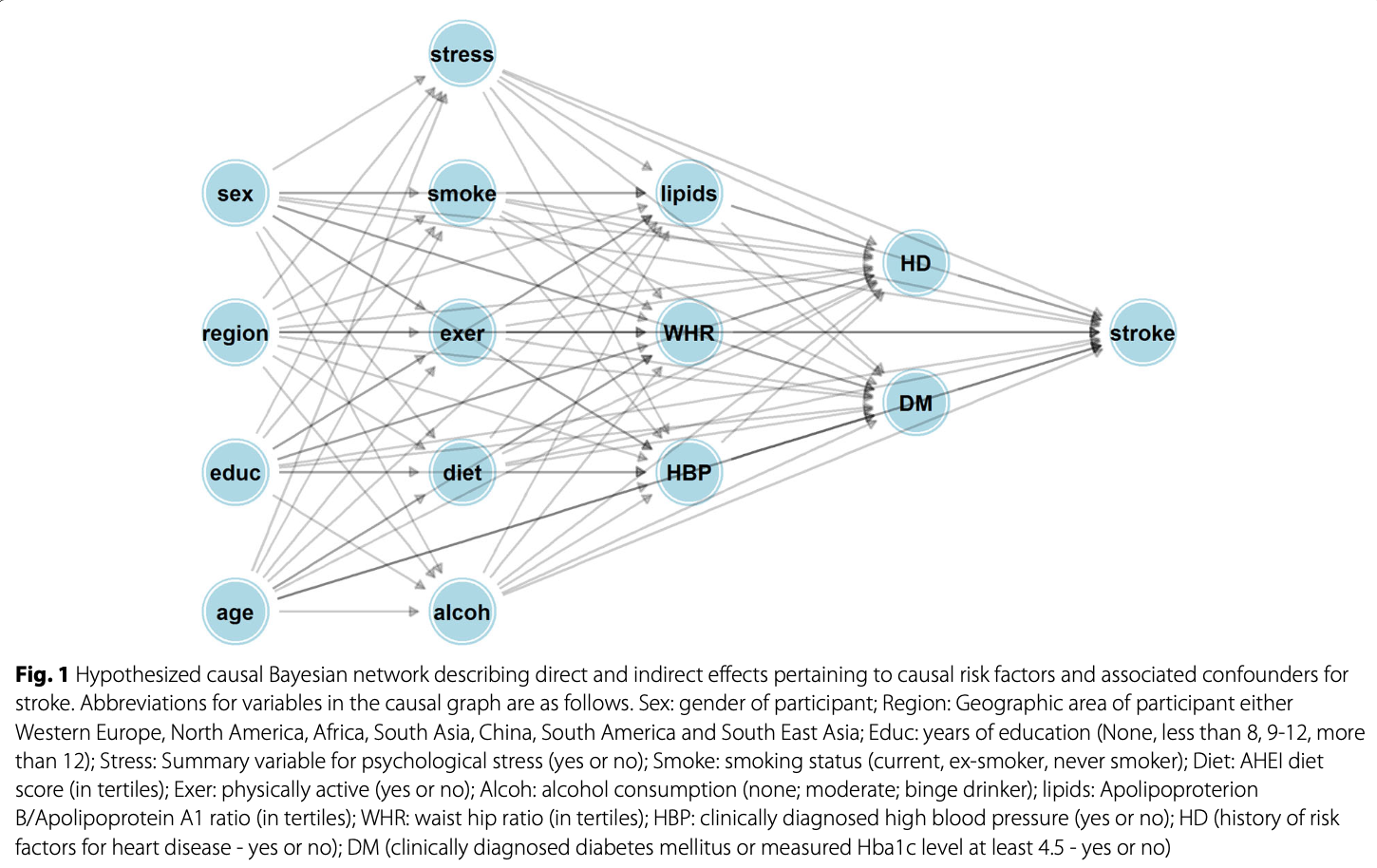 options: width=100 alt="Causal Bayesian network DAG"
options: width=100 alt="Causal Bayesian network DAG"
Usage
indirect_PAF_Sjolander_onesimulation(
data_frame,
exposure,
mediator,
response,
mediator_model,
response_model,
response_model_2,
weights
)
Arguments
data_frame |
Data frame containing the data. The data frame has cases in rows and variables in columns. |
exposure |
The exposure name in the form of character string e.g. “phys” for physical exercise. |
mediator |
The mediator name in the form of character string e.g. “whr” for waist hip ratio. |
response |
The outcome name in the form of character string e.g. “case” for a stroke case. |
mediator_model |
A list containing each of the fitted mediator regression models e.g.
|
response_model |
is a regression for the outcome on all mediators together with all parents and confounders of the mediators in a Markov causal Bayesian network DAG e.g. |
response_model_2 |
is a regression for the outcome on the exposure together with all parents and confounders of the exposure in a Markov causal Bayesian network DAG along with other risk factors at the same level of the causal Bayesian network DAG. E.G. If physical exercise (“exer”) in the example given in the diagram is the exposure. Then the regression would include all parents of “exer” (i.e. sex, region, educ, age) as well as risk factors at the same level of the causal Bayesian network (i.e. stress, smoke, diet, alcoh). |
weights |
A numeric n x |
Value
totalPAF |
total PAF |
directPAF |
direct PAF |
indirectPAF |
indirect PAF |
Creates a DAG for input into package Dagitty to identify adjustmentSets given exposure and outcome
Description
Creates a DAG for input into package Dagitty to identify adjustmentSets given exposure and outcome
Usage
make_DAG(
in_outDAG,
exposure,
response,
mediator,
Splines_outlist_Var = list(),
splinesDefinedIn_in_outDAG = list(),
addCustomExposureAdjustmentSet = FALSE,
customExposureAdjustmentSet,
addCustomMediatorAdjustmentSet = FALSE,
customMediatorAdjustmentSet
)
Arguments
in_outDAG |
This defines the causal directed acyclic graph (DAG). A list of length 2. It is defined as a two dimensional list consisting of, firstly, the first list, inlist, i.e. a list of the parents of each variable of interest corresponding to its column name in the data. Splines can be included here if they are to be modelled as splines. Secondly, the second list, outlist, contains a list of a single name of exposure or risk factor or outcome in form of characters i.e. a list of each variable of interest (risk factors, exposures and outcome) corresponding to its column name in the data. Splines should not be input here, only the column names of the variables of interest in the data. The order at which variables are defined must satisfy (i) It is important that variables are defined in the same order in both lists e.g. the first risk factor defined in outlist has its parents listed first in inlist, the second risk factor defined in outlist has its parents listed secondly in inlist and so on. The package assumes this ordering and will not work if this order is violated. (ii) Note it is important also that the order at which the variables are defined is such that all parents of that variable are defined before it. See example in tutorial. |
exposure |
The name of the exposure column variable within dataframe in text format e.g. “phys”. |
response |
The name of the response column variable within dataframe in text format e.g. “case”. The cases should be coded as 1 and the controls as 0. |
mediator |
The name of the mediator column variables within dataframe in text format. There can be more than one mediator of interest. It can be a vector of mediators names within the dataframe e.g. c(“subhtn”,“apob_apoa”,“whr”). |
Splines_outlist_Var |
A list defined of same size and order of variables as defined in in_outArg[[2]]. If splines are to be used for variables listed in in_outArg[[2]], then the splines should be defined in Splines_outlist in the same order as variables appear in in_outArg[[2]]. It is necessary to list variables in Splines_outlist the same as in in_outArg[[2]] without splines if no spline is to be applied. It should not be input as an empty list, list(), if no splines. A warning will show if input as an empty list requiring the user to populate Splines_outlist either the same as in_outArg[[2]] (if no splines) or in the same order as in_outArg[[2]] with splines (if splines). See example in tutorial. |
splinesDefinedIn_in_outDAG |
Logical TRUE or FALSE indicating whether the user has defined splines in the causal DAG, in_out, if TRUE. If FALSE and splines are defined in Splines_outlist_Var, then it is necessary for the package to populate the in_out DAG with splines listed in Splines_outlist_Var. |
addCustomExposureAdjustmentSet |
Logical TRUE or FALSE indicating whether a customised interaction term is to be added to the each regression for the Exposure adjustment set. The interaction term can include splines. NOTE variables in addCustom are input by the User and the User should decide whether they are to be included in the adjustmentSet and whether these variables are not already present. The DAG will be fit without nodes in addCustom since causal DAG nodes will not recognise customised nodes e.g. with a customised formula in the node name. The addCustom is added in additively as adjustment set variables after the causal diagram has been defined and adjustment sets have been identified. Care should be taken when using addCustom not to add in variables eg. twice if already included as variables in the causal dag. |
customExposureAdjustmentSet |
text containing the customised interaction term to be added to each regression for the Exposure adjustment set. The text should be enclosed in inverted commas. Splines can be included within the interaction terms. If there are more than one exposure and the custom is to differ per exposure, custom can be defined in a vector of form c(), where each customised adjustment is in the same order as the variables defined in exposure if more than one exposure. See tutorial for examples. NOTE variables in addCustom are input by the User and the User should decide whether they are to be included in the adjustmentSet and whether these variables are not already present. The DAG will be fit without nodes in addCustom since causal DAG nodes will not recognise customised nodes e.g. with a customised formula in the node name. The addCustom is added in additively as adjustment set variables after the causal diagram has been defined and adjustment sets have been identified. Care should be taken when using addCustom not to add in variables eg. twice if already included as variables in the causal dag. |
addCustomMediatorAdjustmentSet |
Logical TRUE or FALSE indicating whether a customised interaction term is to be added to the each regression for the Mediator adjustment set. The interaction term can include splines. NOTE variables in addCustom are input by the User and the User should decide whether they are to be included in the adjustmentSet and whether these variables are not already present. The DAG will be fit without nodes in addCustom since causal DAG nodes will not recognise customised nodes e.g. with a customised formula in the node name. The addCustom is added in additively as adjustment set variables after the causal diagram has been defined and adjustment sets have been identified. Care should be taken when using addCustom not to add in variables eg. twice if already included as variables in the causal dag. |
customMediatorAdjustmentSet |
text containing the customised interaction term to be added to each regression for the Mediator adjustment set. The text should be enclosed in inverted commas. Splines can be included within the interaction terms. If there are more than one mediator and the custom is to differ per mediator, custom can be defined in a vector of form c(), where each customised adjustment is in the same order as the variables defined in mediator if more than one mediator. See tutorial for examples. NOTE variables in addCustom are input by the User and the User should decide whether they are to be included in the adjustmentSet and whether these variables are not already present. The DAG will be fit without nodes in addCustom since causal DAG nodes will not recognise customised nodes e.g. with a customised formula in the node name. The addCustom is added in additively as adjustment set variables after the causal diagram has been defined and adjustment sets have been identified. Care should be taken when using addCustom not to add in variables eg. twice if already included as variables in the causal dag. |
Value
Returns DAG and adjustment set given exposure and outcome.
Creates a DAG for input into package Dagitty to identify adjustmentSets given exposure and outcome.
Description
Creates a DAG for input into package Dagitty to identify adjustmentSets given exposure and outcome. Splines are removed initially and then added back in. Adjustment sets populated by the user are updated with valid adjustment sets if necessary.
Usage
make_DAG_AdjustmentSets_in_out(
in_outDAG,
exposure,
response,
mediator,
Splines_outlist_Var = list(),
splinesDefinedIn_in_outDAG = list(),
addCustomExposureAdjustmentSet = FALSE,
customExposureAdjustmentSet,
addCustomMediatorAdjustmentSet = FALSE,
customMediatorAdjustmentSet
)
Arguments
in_outDAG |
This defines the causal directed acyclic graph (DAG). A list of length 2. It is defined as a two dimensional list consisting of, firstly, the first list, inlist, i.e. a list of the parents of each variable of interest corresponding to its column name in the data. Splines can be included here if they are to be modelled as splines. Secondly, the second list, outlist, contains a list of a single name of exposure or risk factor or outcome in form of characters i.e. a list of each variable of interest (risk factors, exposures and outcome) corresponding to its column name in the data. Splines should not be input here, only the column names of the variables of interest in the data. The order at which variables are defined must satisfy (i) It is important that variables are defined in the same order in both lists e.g. the first risk factor defined in outlist has its parents listed first in inlist, the second risk factor defined in outlist has its parents listed secondly in inlist and so on. The package assumes this ordering and will not work if this order is violated. (ii) Note it is important also that the order at which the variables are defined is such that all parents of that variable are defined before it. See example in tutorial. |
exposure |
The name of the exposure column variable within dataframe in text format e.g. “phys”. |
response |
The name of the response column variable within dataframe in text format e.g. “case”. The cases should be coded as 1 and the controls as 0. |
mediator |
The name of the mediator column variables within dataframe in text format. There can be more than one mediator of interest. It can be a vector of mediators names within the dataframe e.g. c(“subhtn”,“apob_apoa”,“whr”). |
Splines_outlist_Var |
A list defined of same size and order of variables as defined in in_outArg[[2]]. If splines are to be used for variables listed in in_outArg[[2]], then the splines should be defined in Splines_outlist in the same order as variables appear in in_outArg[[2]]. It is necessary to list variables in Splines_outlist the same as in in_outArg[[2]] without splines if no spline is to be applied. It should not be input as an empty list, list(), if no splines. A warning will show if input as an empty list requiring the user to populate Splines_outlist either the same as in_outArg[[2]] (if no splines) or in the same order as in_outArg[[2]] with splines (if splines). See example in tutorial. |
splinesDefinedIn_in_outDAG |
Logical TRUE or FALSE indicating whether the user has defined splines in the causal DAG, in_out, if TRUE. If FALSE and splines are defined in Splines_outlist_Var, then it is necessary for the package to populate the in_out DAG with splines listed in Splines_outlist_Var. |
addCustomExposureAdjustmentSet |
Logical TRUE or FALSE indicating whether a customised interaction term is to be added to the each regression for the Exposure adjustment set. The interaction term can include splines. NOTE variables in addCustom are input by the User and the User should decide whether they are to be included in the adjustmentSet and whether these variables are not already present. The DAG will be fit without nodes in addCustom since causal DAG nodes will not recognise customised nodes e.g. with a customised formula in the node name. The addCustom is added in additively as adjustment set variables after the causal diagram has been defined and adjustment sets have been identified. Care should be taken when using addCustom not to add in variables eg. twice if already included as variables in the causal dag. |
customExposureAdjustmentSet |
text containing the customised interaction term to be added to each regression for the Exposure adjustment set. The text should be enclosed in inverted commas. Splines can be included within the interaction terms. If there are more than one exposure and the custom is to differ per exposure, custom can be defined in a vector of form c(), where each customised adjustment is in the same order as the variables defined in exposure if more than one exposure. See tutorial for examples. NOTE variables in addCustom are input by the User and the User should decide whether they are to be included in the adjustmentSet and whether these variables are not already present. The DAG will be fit without nodes in addCustom since causal DAG nodes will not recognise customised nodes e.g. with a customised formula in the node name. The addCustom is added in additively as adjustment set variables after the causal diagram has been defined and adjustment sets have been identified. Care should be taken when using addCustom not to add in variables eg. twice if already included as variables in the causal dag. |
addCustomMediatorAdjustmentSet |
Logical TRUE or FALSE indicating whether a customised interaction term is to be added to the each regression for the Mediator adjustment set. The interaction term can include splines. NOTE variables in addCustom are input by the User and the User should decide whether they are to be included in the adjustmentSet and whether these variables are not already present. The DAG will be fit without nodes in addCustom since causal DAG nodes will not recognise customised nodes e.g. with a customised formula in the node name. The addCustom is added in additively as adjustment set variables after the causal diagram has been defined and adjustment sets have been identified. Care should be taken when using addCustom not to add in variables eg. twice if already included as variables in the causal dag. |
customMediatorAdjustmentSet |
text containing the customised interaction term to be added to each regression for the Mediator adjustment set. The text should be enclosed in inverted commas. Splines can be included within the interaction terms. If there are more than one mediator and the custom is to differ per mediator, custom can be defined in a vector of form c(), where each customised adjustment is in the same order as the variables defined in mediator if more than one mediator. See tutorial for examples. NOTE variables in addCustom are input by the User and the User should decide whether they are to be included in the adjustmentSet and whether these variables are not already present. The DAG will be fit without nodes in addCustom since causal DAG nodes will not recognise customised nodes e.g. with a customised formula in the node name. The addCustom is added in additively as adjustment set variables after the causal diagram has been defined and adjustment sets have been identified. Care should be taken when using addCustom not to add in variables eg. twice if already included as variables in the causal dag. |
Value
Returns DAG and adjustment set given exposure and outcome with spline variables included and updates any adjustment sets if necessary for valid adjustments.
Makes Formula for regression of exposure or risk factor or outcome on its parents in a causal Bayesian network directed acyclic graph.
Description
Given a causal Bayesian network, directed acyclic graph (DAG) where arrows representing
causal dependencies between confounders, risk factors, exposure and disease, together with a sensible probability distribution on
the graph that respects these causal dependencies. To consistently estimate causal effects that risk factors may have on each
other and on disease, we need to make a strong no unmeasured confounding assumption: that is common causes of nodes in the graph,
which may be causes of two risk factors or a cause of risk factor and disease, are also included as nodes in the graph.
Causal Bayesian networks have a local Markov property that the conditional probability distribution of any node X_j, given values
for the other variables in the network, only depends on the values x_{pa_{j}} of the parent nodes.
Usage
make_formula(in_vars, outvar, addCustom = FALSE, custom = "")
Arguments
in_vars |
a list of character vectors of the parents of the exposure or risk factor or outcome which are either causes or confounders of the exposure or risk factor or outcome |
outvar |
list of a single name of exposure or risk factor or outcome in form of characters |
addCustom |
is a logical which is TRUE if a custom regression is to be added in or FALSE otherwise. The customised part is defined in the parameter “custom”. |
custom |
custom must start and end as follows: "~ + " with custom values between ~ and +. Custom allows customised input into regression not stated already e.g. allow for interaction of variables not listed yet e.g. "~ regionnn7 x ns(eage,df=5)+esex x ns(eage,df=5) + " |
Value
result |
result |
Calculation of Population Attributable Fraction (PAF), with a decomposition of the total PAF into direct and indirect components.
Description
Total PAF
Usage
overall_direct(
data_frame,
exposure,
mediator,
response,
mediator_model,
response_model,
response_model_2,
weights
)
Arguments
data_frame |
Data frame containing the data. The data frame has cases in rows and variables in columns. |
exposure |
The exposure name in the form of character string e.g. "phys" for physical exercise. |
mediator |
The mediator name in the form of character string e.g. "whr" for waist hip ratio. |
response |
The outcome name in the form of character string e.g. "case" for a stroke case. |
mediator_model |
A list containing each of the mediator regression models e.g. |
response_model |
is a regression model fitted for the outcome on all mediators together with all parents and confounders of the mediators in a Markov causal Bayesian network DAG. |
response_model_2 |
A regression model fitted for the response in a causal Bayesian network excluding “children” of the exposure in the causal Bayesian network. This regression model will not adjust for mediators (exclude mediators) of the exposure in the regression model so that the total effect of the exposure on the response can be modelled. This model can be listed either as (1) an empty list ( response_model_exposure = list() ) or (2) the user can specify their own customised causal regression model to use. If specified as an empty list, list(), then the 'causalPAF' package will define and fit the model automatically based on the causal DAG defined by the in_outArg parameter. Alternatively, the user can specify the exact model that the user wishes to use, this model must be in list format (list() where length(response_model_exposure) == 1 ), of length 1, assuming only one exposure of interest (other exposures can be risk factors) and the model must be defined within a list() since the package assumes a list() format is supplied. See example in tutorial. E.G. If physical exercise ("exer") in the example given in the diagram is the exposure. Then the regression would include all parents of "exer" (i.e. sex, region, educ, age) as well as risk factors at the same level of the causal Bayesian network (i.e. stress, smoke, diet, alcoh). |
weights |
A numeric |
Value
directPAF |
direct PAF |
Path specific population attributable fraction for a mediating pathway.
Description
Path specific population attributable fraction for a mediating pathway. This metric has several desirable properties. This is a kind of sequential PAF on pathways, but now corresponding to eliminating the mediating pathway first.
PAF_{A->M->Y} = (P(Y=1)-P(Y_{A,M_{0}}=1)/P(Y=1)
 options: width=100 alt="Causal Bayesian network DAG"
options: width=100 alt="Causal Bayesian network DAG"
Usage
path_specific_onesimulation(
data_frame,
exposure,
mediator,
response,
mediator_model,
response_model,
response_model_2,
weights
)
Arguments
data_frame |
Data frame containing the data. The data frame has cases in rows and variables in columns. |
exposure |
The exposure name in the form of character string e.g. "phys" for physical exercise. |
mediator |
The mediator name in the form of character string e.g. "whr" for waist hip ratio. |
response |
The outcome name in the form of character string e.g. "case" for a stroke case. |
mediator_model |
A list containing each of the fitted mediator regression models e.g.
|
response_model |
is a regression for the outcome on all mediators together with all parents and confounders of the mediators in a Markov causal Bayesian network DAG. A regression model fitted for the response in a causal Bayesian network excluding “children” of the mediators in the causal Bayesian network. See example in tutorial.This model can be listed either as (1) an empty list ( response_model_mediators = list() ) or (2) the user can specify their own customised causal regression model(s) to use. When it is listed as an empty list the 'causalPAF' package will fit the response_model_mediators regression model automatically based on the causal DAG supplied by the user in in_outArg. Alternatively, the user can specify the exact model(s) that the user wishes to use, these model(s) must be in list format (list() where length(response_model_mediators) == length(mediator) ), the same length as the parameter, mediator, with the user customised model for each mediator listed in the same order as in the parameter, mediator, and if there is only one model, it must be listed each time within the list() so that length(response_model_mediators) == length(mediator). |
response_model_2 |
A regression model fitted for the response in a causal Bayesian network excluding “children” of the exposure in the causal Bayesian network. This regression model will not adjust for mediators (exclude mediators) of the exposure in the regression model so that the total effect of the exposure on the response can be modelled. This model can be listed either as (1) an empty list ( response_model_exposure = list() ) or (2) the user can specify their own customised causal regression model to use. If specified as an empty list, list(), then the 'causalPAF' package will define and fit the model automatically based on the causal DAG defined by the in_outArg parameter. Alternatively, the user can specify the exact model that the user wishes to use, this model must be in list format (list() where length(response_model_exposure) == 1 ), of length 1, assuming only one exposure of interest (other exposures can be risk factors) and the model must be defined within a list() since the package assumes a list() format is supplied. See example in tutorial. E.G. If physical exercise ("exer") in the example given in the diagram is the exposure. Then the regression would include all parents of "exer" (i.e. sex, region, educ, age) as well as risk factors at the same level of the causal Bayesian network (i.e. stress, smoke, diet, alcoh). |
weights |
A numeric n x |
Value
path_specific_PAF |
path specific PAF |
Evaluates Point Estimates for Total PAF, Direct PAF, Indirect PAF and Path Specific PAF for a user inputted number of integral simulations. There is no bootstrap applied in this function.
Description
Evaluates Total PAF, Direct PAF, Indirect PAF and Path Specific PAF for a user inputted number of bootstraps and integral simulations
Usage
pointEstimate(
dataframe,
exposure = "phys",
mediator = c("subhtn", "apob_apoa", "whr"),
response = "case",
response_model_mediators = list(),
response_model_exposure = list(),
in_outArg,
Splines_outlist,
splinesDefinedIn_in_outDAG,
model_listArg,
weights = 1,
NumSimulation,
addCustom = FALSE,
custom = ""
)
Arguments
dataframe |
A wide format dataframe containing all the risk factors, confounders, exposures and outcomes within the causal DAG Bayesian network. |
exposure |
The name of the exposure column variable within dataframe in text format e.g. "phys". |
mediator |
The name of the mediator column variables within dataframe in text format. There can be more than one mediator of interest. It can be a vector of mediators names within the dataframe e.g. c("subhtn","apob_apoa","whr"). |
response |
The name of the response column variable within dataframe in text format e.g. "case". The cases should be coded as 1 and the controls as 0. |
response_model_mediators |
A regression model fitted for the response in a causal Bayesian network excluding “children” of the mediators in the causal Bayesian network. See example in tutorial.This model can be listed either as (1) an empty list ( response_model_mediators = list() ) or (2) the user can specify their own customised causal regression model(s) to use. When it is listed as an empty list the 'causalPAF' package will fit the response_model_mediators regression model automatically based on the causal DAG supplied by the user in in_outArg. Alternatively, the user can specify the exact model(s) that the user wishes to use, these model(s) must be in list format (list() where length(response_model_mediators) == length(mediator) ), the same length as the parameter, mediator, with the user customised model for each mediator listed in the same order as in the parameter, mediator, and if there is only one model, it must be listed each time within the list() so that length(response_model_mediators) == length(mediator). |
response_model_exposure |
A regression model fitted for the response in a causal Bayesian network excluding “children” of the exposure in the causal Bayesian network. This regression model will not adjust for mediators (exclude mediators) of the exposure in the regression model so that the total effect of the exposure on the response can be modelled. This model can be listed either as (1) an empty list ( response_model_exposure = list() ) or (2) the user can specify their own customised causal regression model to use. If specified as an empty list, list(), then the 'causalPAF' package will define and fit the model automatically based on the causal DAG defined by the in_outArg parameter. Alternatively, the user can specify the exact model that the user wishes to use, this model must be in list format (list() where length(response_model_exposure) == 1 ), of length 1, assuming only one exposure of interest (other exposures can be risk factors) and the model must be defined within a list() since the package assumes a list() format is supplied. See example in tutorial. E.G. If physical exercise ("exer") in the example given in the diagram is the exposure. Then the regression would include all parents of "exer" (i.e. sex, region, educ, age) as well as risk factors at the same level of the causal Bayesian network (i.e. stress, smoke, diet, alcoh). |
in_outArg |
This defines the causal directed acyclic graph (DAG). A list of length 2. It is defined as a two dimensional list consisting of, firstly, the first list, inlist, i.e. a list of the parents of each variable of interest corresponding to its column name in the data. Splines can be included here if they are to be modelled as splines. Secondly, the second list, outlist, contains a list of a single name of exposure or risk factor or outcome in form of characters i.e. a list of each variable of interest (risk factors, exposures and outcome) corresponding to its column name in the data. Splines should not be input here, only the column names of the variables of interest in the data. The order at which variables are defined must satisfy (i) It is important that variables are defined in the same order in both lists e.g. the first risk factor defined in outlist has its parents listed first in inlist, the second risk factor defined in outlist has its parents listed secondly in inlist and so on. The package assumes this ordering and will not work if this order is violated. (ii) Note it is important also that the order at which the variables are defined is such that all parents of that variable are defined before it. See example in tutorial. |
Splines_outlist |
A list defined of same size and order of variables as defined in in_outArg[[2]]. If splines are to be used for variables listed in in_outArg[[2]], then the splines should be defined in Splines_outlist in the same order as variables appear in in_outArg[[2]]. It is necessary to list variables in Splines_outlist the same as in in_outArg[[2]] without splines if no spline is to be applied. It should not be input as an empty list, list(), if no splines. A warning will show if input as an empty list requiring the user to populate Splines_outlist either the same as in_outArg[[2]] (if no splines) or in the same order as in_outArg[[2]] with splines (if splines). See example in tutorial. |
splinesDefinedIn_in_outDAG |
Logical TRUE or FALSE indicating whether the user has defined splines in the causal DAG, in_out, if TRUE. If FALSE and splines are defined in Splines_outlist_Var, then it is necessary for the package to populate the in_out DAG with splines listed in Splines_outlist_Var. |
model_listArg |
is a list of models fitted for each of the variables in in_outArg[[2]] (or in_outArg |
weights |
Column of weights for case control matching listed in the same order as the patients in the data e.g. weights = strokedata$weights. |
NumSimulation |
This is the number of simulations requested by the user to estimate integrals. The larger the number of simulations the more accurate the results but the longer the code takes to run. Therefore the user may wish to balance speed with accuracy depending on which is of more value in the specific context of interest. The integrals for continuous variables are estimated using simulation methods. |
addCustom |
Logical TRUE or FALSE indicating whether a customised interaction term is to be added to the each regression. The interaction term can include splines. |
custom |
text containing the customised interaction term to be added to each regression. The text should be enclosed in inverted commas. Splines can be included within the interaction terms. See tutorial for examples. |
Value
Estimates point estimates for 5 results that are:(1)Total Population Attributable Fraction (PAF),(2)Direct Effect Population Attributable Fraction (PAF) using alternative definition, (3)Indirect Effect Population Attributable Fraction (PAF) using alternative definition, (4)Path Specific Population Attributable Fraction (PAF), (5)Overall Direct Population Attributable Fraction (PAF)
Examples
# Loads some data (fictional Stroke data from the package 'causalPAF')
# In this example, we use a small data set called 'strokedata_smallSample' consisting of 5,000
# rows of fictional patient data. For more accurate results, a larger data set is available
# called 'strokedata'which contains 16,623 rows of fictional patient data. The methodology
# applied in the 'causalPAF' package is more accurate the larger the dataset. To use the larger
# 'strokedata' dataset, simply call
# stroke_reduced <- strokedata
stroke_reduced <- strokedata_smallSample
# Just shortening the name of a variable, "apob_apoa", to "apb" so the R code
# in document example is not truncated.
stroke_reduced$apb <- stroke_reduced$apob_apoa
# The data should contain a column of weights for case control matching.
# strokedata$weights
# Weigths are not needed for cohort/cross sectional designs.
# The data should have reference levels of all risk factors already set.
# This can be done as follows but has already been applied to the data so is not run here:
# levels(stroke_reduced$htnadmbp) <- c(0, 1)
# stroke_reduced$subhtn <- factor(stroke_reduced$subhtn,levels=c(1, 2))
# levels(stroke_reduced$nevfcur) <- c(1, 2)
# stroke_reduced$global_stress2 <- factor(stroke_reduced$global_stress2,levels=c(1,2))
# levels(stroke_reduced$whrs2tert) <- c(1, 2, 3)
# levels(stroke_reduced$phys) <- c(2, 1)
# levels(stroke_reduced$alcohfreqwk) <- c(1, 2, 3)
# stroke_reduced$dmhba1c2 <- factor(stroke_reduced$dmhba1c2,levels=c(1,2))
# stroke_reduced$cardiacrfcat <- factor(stroke_reduced$cardiacrfcat,levels=c(1,2))
# levels(stroke_reduced$ahei3tert) <- c(3,2,1)
# levels(stroke_reduced$apob_apoatert) <- c(1,2,3)
# The 'causalPAF' package assumes the data is either complete case data or that missing data
# analysis has already been performed.
# Next, define the causal structure or directed acyclic graph (DAG) of the causal Bayesian
# network defined by the data. We list the parents of each exposure or risk factor or outcome
# in a vector as follows:
# Note it is important that the order at which the variables are defined is such that all
# parents of that variable are defined before it. Please refer to the figure of the causal
# Bayesian network (with both direct and indirect effects) defined earlier as an example of this
# order.
in_phys <- c("subeduc","moteduc","fatduc")
in_ahei <- c("subeduc","moteduc","fatduc")
in_nevfcur <- c("subeduc","moteduc","fatduc")
in_alcohfreqwk <- c("subeduc","moteduc","fatduc")
in_global_stress2 <- c("subeduc","moteduc","fatduc")
in_subhtn <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_apob_apoa <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_whr <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
# Note splines can be fitted within the causal structure as shown below especially if splines
# are to be used in the fitted models.
# It is important that splines of parent variables are "typed" or "spelt" consistently
# (including spaces) throughout as 'causalPAF' can fit models automatically provided variables are
# spelt consistently. Also if a parent variable is a spline it should be defined in spline
# format in all occurences of the parent variable.
in_cardiacrfcat <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2",
"ns(apb,knots=quantile(apb,c(.25,.5,.75)),Boundary.knots=quantile(apb,c(.001,.95)))",
"ns(whr,df=5)","subhtn")
in_dmhba1c2 <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2",
"ns(apb,knots=quantile(apb,c(.25,.5,.75)),Boundary.knots=quantile(apb,c(.001,.95)))",
"ns(whr,df=5)","subhtn")
in_case <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2",
"ns(apb,knots=quantile(apb,c(.25,.5,.75)),Boundary.knots=quantile(apb,c(.001,.95)))",
"ns(whr,df=5)","subhtn","cardiacrfcat","dmhba1c2")
# Then we define a two dimensional list consisting of
# 1. inlist i.e. a list of the parents of each variable of interest corresponding to its column
# name in the data. Splines should be included here if they are to be modelled as splines.
# 2. outlist i.e. a list of each variable of interest corresponding to its column name in the
# data. Splines should not be input here, only the column names of the variables of interest in
# the data.
# Again the order is such that each variable is defined after all its parents.
in_out <- list(inlist=list(in_phys,in_ahei,in_nevfcur,in_alcohfreqwk,in_global_stress2,
in_subhtn,in_apob_apoa,in_whr,in_cardiacrfcat,in_dmhba1c2,in_case),
outlist=c("phys","ahei3tert","nevfcur","alcohfreqwk","global_stress2","subhtn",
"apb","whr","cardiacrfcat","dmhba1c2","case"))
# If splines are to be used for variables listed in in_out$outlist, then the splines should be
# defined in the same order as variables appear in in_out$outlist as follows. It is necessary to
# list variables in in_out$outlist without splines if no spline is to be applied.
# It is important that Splines_outlist is defined in the following format
# list(c("splinename1","splinename2","splinename3")) for the package to be applied correctly.
# And Splines_outlist should not be an empty list(). If there are no splines it should be
# defined the same as in_out[[2]] and in the same order as variables defined in_out[[2]].
Splines_outlist = list( c("phys","ahei3tert","nevfcur","alcohfreqwk","global_stress2","subhtn",
"ns(apb,knots=quantile(apb,c(.25,.5,.75)),Boundary.knots=quantile(apb,c(.001,.95)))",
"ns(whr,df=5)","cardiacrfcat","dmhba1c2","case") )
# To fit these models to case control data, one needs to perform weighted maximum likelihood
# estimation to imitate estimation using a random sample from the population. We chose weights
# of 0.0035 (for each case) and 0.9965 (for each control), reflective of a yearly incidence of
# first ischemic stroke of 0.35%, or 3.5 strokes per 1,000 individuals. These weights were
# chosen according to average incidences across country, age, group and gender within
# INTERSTROKE according to the global burden of disease.
w <- rep(1,nrow(stroke_reduced))
w[stroke_reduced$case==0] <- 0.9965
w[stroke_reduced$case==1] <- 0.0035
# It is important to assign stroke_reduced$weights to the updated weights defined in w.
# Otherwise if stroke_reduced$weights <- w is not set, the alternative weights supplied in the
# fictional data will be used. In this case, we want to use weigths as defined in w.
stroke_reduced$weights <- w
#The checkMarkovDAG() function in the 'causalPAF' package should be used before running
# causalPAFplot() to ensure:
#1. The causal Markov condition holds for the causal structure defined in the variable in_out.
#2. The variables in in_out are listed in the order so that no variable is defined before a
# parent or direct cause. Note: if this order does not hold, checkMarkovDAG() will automatically
# reorder the variables in, in_out, provided it is a Markov DAG.
#The causal analysis requires that the causal structure is a Markov DAG. The Causal Markov (CM)
# condition states that, conditional on the set of all its direct causes, a node is independent
# of all variables which are not direct causes or direct effects of that node. In the event that
# the structure of a Bayesian network accurately depicts causality, the two conditions are
# equivalent. However, a network may accurately embody the Markov condition without depicting
# causality, in which case it should not be assumed to embody the causal Markov condition.
# in_out is as defined above and input into this code.
if(checkMarkovDAG(in_out)$IsMarkovDAG & !checkMarkovDAG(in_out)$Reordered){
print("Your in_out DAG is a Markov DAG.")
} else if( checkMarkovDAG(in_out)$IsMarkovDAG & checkMarkovDAG(in_out)$Reordered ) {
in_out <- checkMarkovDAG(in_out)[[2]]
print("Your in_out DAG is a Markov DAG.The checkMarkovDAG function has reordered your
in_out list so that all parent variables come before descendants.")
} else{ print("Your ``in_out'' list is not a Bayesian Markov DAG so the methods in the
'causalPAF' package cannot be applied for non Markov DAGs.")}
# The pointEstimate() function evaluates Point Estimates for Total PAF, Direct PAF, Indirect PAF
# and Path Specific PAF for a user inputted number of integral simulations. There is no bootstrap
# applied in this function.
# Since bootstraps are not applied, the pointEstimate() function will run quicker than the
# alternative causalPAFplot() function which calculates bootstrap estimates which can take
# longer to run.
pointEstimate(dataframe = stroke_reduced,
exposure="phys",
mediator=c("subhtn","apb","whr"),
response="case",
response_model_mediators = list(),
response_model_exposure = list(),
in_outArg = in_out,
Splines_outlist = Splines_outlist,
splinesDefinedIn_in_outDAG = TRUE,
model_listArg = list(),
weights = w,
NumSimulation = 3,
addCustom = TRUE,
custom = "regionnn7*ns(eage,df=5)+esex*ns(eage,df=5)")
# The pointEstimate() function below has response_model_mediators, response_model_exposure and
# model_listArg pre fit. This allows the user to apply customised regressions instead of the
# default setting above, where the 'causalPAF' R package fitted these regressions automatically
# based on the causalDAG defined in in_outArg.
# Libraries must be loaded if fitting models outside of the 'causalPAF' R package.
library(MASS)
library(splines)
# Next we fit the, response_model_mediators and response_model_exposure, models outside of the
# 'causalPAF' package as an input into the package.
# It is important that response_vs_mediator is a list and it must be the same length as the
# parameter, mediator, i.e. length( response_vs_mediator ) == length( mediator). In this
# example, mediator=c("subhtn","apb","whr") so length( mediator) is 3, so we create a list
# with three models for "subhtn","apb" and "whr" respectively in that order. Note in this
# example, the model is the same for each mediator, but it must still be input 3 times within
# the list as follows:
response_vs_mediator <- list(
glm("case ~ regionnn7*ns(eage,df=5)+esex*ns(eage,df=5) +subeduc+moteduc+ fatduc+ phys+
ahei3tert+ nevfcur+ alcohfreqwk+ global_stress2+ subhtn +
ns(apb, knots = quantile(apb,c(.25,0.5,0.75)),
Boundary.knots = quantile(apb,c(.001,0.95)))+
ns(whr,df=5)",data = stroke_reduced,family='binomial',w = stroke_reduced$weights ),
# "subhtn" mediator model
glm("case ~ regionnn7*ns(eage,df=5)+esex*ns(eage,df=5) + subeduc+ moteduc+ fatduc+ phys+
ahei3tert+ nevfcur+ alcohfreqwk+ global_stress2+ subhtn +
ns(apb, knots = quantile(apb,c(.25,0.5,0.75)),
Boundary.knots = quantile(apb,c(.001,0.95)))+
ns(whr,df=5)",data = stroke_reduced,family='binomial',w = stroke_reduced$weights ),
# "apb" mediator model
glm("case ~ regionnn7*ns(eage,df=5)+esex*ns(eage,df=5) + subeduc+ moteduc+ fatduc+ phys+
ahei3tert+ nevfcur+ alcohfreqwk+ global_stress2+ subhtn +
ns(apb, knots = quantile(apb,c(.25,0.5,0.75)),
Boundary.knots = quantile(apb,c(.001,0.95)))+
ns(whr,df=5)",data = stroke_reduced,family='binomial',w = stroke_reduced$weights ) )
# "whr" mediator model
# Next we fit a customised response_model_exposure model rather than allowing the package fit it
# automatically as shown previously. This must be a list of length 1.
response_vs_phys <- list(glm("case ~ regionnn7*ns(eage,df=5)+esex*ns(eage,df=5)+subeduc+moteduc+
fatduc+ phys+ ahei3tert+ nevfcur+ alcohfreqwk+ global_stress2",data = stroke_reduced,
family='binomial',w= stroke_reduced$weights) )
# model_listArg is a list of models fitted for each of the variables in in_out$outlist based on
# its parents given in in_out$inlist. By default this is set to an empty list. Alternatively the
# user can supply their custom fitted, model_listpop, which should be consistent with the causal
# structure. model_listArg is defined earlier in this tutorial.
# Note it is important that model_listArg is defined as a list and in the same order and length
# as the variables defined in in_outArg[[2]].
model_listArgFit <- list(glm(formula = phys ~ subeduc + regionnn7 * ns(eage, df = 5) +
esex * ns(eage, df = 5) + moteduc + fatduc, family = "binomial", data = stroke_reduced,
weights = weights), # model 1 phys
polr(formula = ahei3tert ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, data = stroke_reduced, weights = weights), # model 2 ahei3tert
glm(formula = nevfcur ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, family = "binomial",data = stroke_reduced, weights = weights),
# model 3 nevfcur
polr(formula = alcohfreqwk ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc, data = stroke_reduced,weights = weights), # model 4 alcohfreqwk
glm(formula = global_stress2 ~ subeduc + regionnn7 * ns(eage,df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, family = "binomial",data = stroke_reduced, weights = weights),
# model 5 global_stress2
glm(formula = subhtn ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2,family = "binomial",
data = stroke_reduced, weights = weights), # model 6 subhtn
lm(formula = apb ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2,
data = stroke_reduced,weights = weights), # model 7 apob_apoa coded as apb
lm(formula = whr ~ subeduc + regionnn7 * ns(eage, df = 5) + esex *ns(eage, df = 5) + moteduc +
fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2, data = stroke_reduced,
weights = weights), # model 8 whr
glm(formula = cardiacrfcat ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2 +
ns(apb, knots = quantile(apb,c(0.25, 0.5, 0.75)),
Boundary.knots = quantile(apb,c(0.001, 0.95))) + ns(whr, df = 5) + subhtn,
family = "binomial",data = stroke_reduced, weights = weights), # model 9 cardiacrfcat
glm(formula = dmhba1c2 ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2 +
ns(apb, knots = quantile(apb,c(0.25, 0.5, 0.75)),
Boundary.knots = quantile(apb,c(0.001, 0.95))) + ns(whr, df = 5) + subhtn,
family = "binomial",data = stroke_reduced, weights = weights), # model 10 dmhba1c2
glm(formula = case ~ subeduc + regionnn7 * ns(eage, df = 5) +esex * ns(eage, df = 5) + moteduc +
fatduc + phys + ahei3tert +nevfcur + alcohfreqwk + global_stress2 +
ns(apb, knots = quantile(apb,c(0.25, 0.5, 0.75)),
Boundary.knots = quantile(apb,c(0.001, 0.95))) + ns(whr, df = 5) + subhtn +
cardiacrfcat +dmhba1c2, family = "binomial", data = stroke_reduced, weights = weights)
# model 11 case
)
pointEstimate(dataframe = stroke_reduced,
exposure="phys",
mediator=c("subhtn","apb","whr"),
response="case",
response_model_mediators = response_vs_mediator,
response_model_exposure = response_vs_phys,
in_outArg = in_out,
Splines_outlist = Splines_outlist,
splinesDefinedIn_in_outDAG = TRUE,
model_listArg = model_listArgFit,
weights = w,
NumSimulation = 3,
addCustom = TRUE,
custom = "regionnn7*ns(eage,df=5)+esex*ns(eage,df=5)")
# See examples from the function causalPAFplot() to see how bootstraps and confidence intervals
# can be calculated in addition to the pont estimates calculated from the pointEstimate()
# function. The causalPAFplot() function also shows examples of plotting these point estimate
# results with confidence intervals.
Sequential Population Attributable Fractions in a Bayesian Network.
Description
'sequential_PAF' calculates and plots sequential population attributable fractions (PAF) under a Bayesian network structure.
Usage
sequential_PAF(
dataframe,
model_list_var,
weights = 1,
in_outDAG,
response,
NumOrderRiskFactors,
addCustom = FALSE,
custom = ""
)
Arguments
dataframe |
A wide format dataframe containing all the risk factors, confounders, exposures and outcomes within the causal DAG Bayesian network. |
model_list_var |
is a list of models fitted for each of the variables in in_outDAG$outlist based on its parents given in in_outDAG$inlist. By default this is set to an empty list. In the default setting, the models are fitted based on the order of the variables input in the parameter in_outArg. See the tutorial for more examples. Alternatively, the user can supply their own fitted models here by populating “model_list_var” with their own fitted models for each risk factor, mediator, exposure and response variable. But the order of these models must be in the same order of the variables in the second list of in_outDAG. See tutorial for further examples. |
weights |
Column of weights for case control matching listed in the same order as the patients in the data e.g. from tutorial weights = strokedata$weights. |
in_outDAG |
This defines the causal directed acyclic graph (DAG). A list of length 2. It is defined as a two dimensional list consisting of, firstly, the first list, inlist, i.e. a list of the parents of each variable of interest corresponding to its column name in the data. Splines can be included here if they are to be modelled as splines. Secondly, the second list, outlist, contains a list of a single name of exposure or risk factor or outcome in form of characters i.e. a list of each variable of interest (risk factors, exposures and outcome) corresponding to its column name in the data. Splines should not be input here, only the column names of the variables of interest in the data. The order at which variables are defined must satisfy (i) It is important that variables are defined in the same order in both lists e.g. the first risk factor defined in outlist has its parents listed first in inlist, the second risk factor defined in outlist has its parents listed secondly in inlist and so on. The package assumes this ordering and will not work if this order is violated. (ii) Note it is important also that the order at which the variables are defined is such that all parents of that variable are defined before it. See example in tutorial. |
response |
The name of the response column variable within dataframe in text format e.g. "case". The cases should be coded as 1 and the controls as 0. |
NumOrderRiskFactors |
is the number of randomly sampled elimination orders (or random permutations of all the risk factors) computing Monte Carlo sequential attributable fractions for each random permutation. |
addCustom |
Logical TRUE or FALSE indicating whether a customised interaction term is to be added to the each regression. The interaction term can include splines. |
custom |
text containing the customised interaction term to be added to each regression. The text should be enclosed in inverted commas. Splines can be included within the interaction terms. See tutorial for examples. |
Details
Patients are listed in rows with variables (i.e. exposure, risk factors, confounders, outcome) listed in columns.
Value
plot |
Returns a plot showing the estimated sequential attributable fractions, by position in elimination order. 95 percent confidence intervals are plotted so that we can be 95 percent confident the true estimate (that would be calculated from the procedure when the number of simulations m approaches infinity) lies in the Monte Carlo interval around the point estimate. The estimates shaded red correspond to a Bayesian network with indirect effects, whereas the estimates shaded blue correspond to the Bayesian network with no indirect effects modelled. Note that the Monte Carlo error at position k incorporates variation due to random selection of the set of risk factors/exposures that are intervened on in stages 1,...k minus 1, and also variation based on the recursive simulation of the disease response. |
SAF_summary |
Returns the data used for the plot containing both the Bayesian network (labelled Bayesian network) with indirect effects modelled and a model (labelled usual) with no indirect effects modelled. |
Examples
# Loads some data (fictional Stroke data from the package 'causalPAF')
# In this example, we use a small data set called 'strokedata_smallSample' consisting of 5,000
# rows of fictional patient data. For more accurate results, a larger data set is available
# called 'strokedata'which contains 16,623 rows of fictional patient data. The methodology
# applied in the 'causalPAF' package is more accurate the larger the dataset. To use the larger
# 'strokedata' dataset, simply call
# stroke_reduced <- strokedata
stroke_reduced <- strokedata_smallSample
in_phys <- c("subeduc","moteduc","fatduc")
in_ahei <- c("subeduc","moteduc","fatduc")
in_nevfcur <- c("subeduc","moteduc","fatduc")
in_alcohfreqwk <- c("subeduc","moteduc","fatduc")
in_global_stress2 <- c("subeduc","moteduc","fatduc")
in_htnadmbp <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_apob_apoatert <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_whrs2tert <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2")
in_cardiacrfcat <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2", "apob_apoatert","whrs2tert","htnadmbp")
in_dmhba1c2 <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2", "apob_apoatert","whrs2tert","htnadmbp")
in_case <- c("subeduc","moteduc","fatduc","phys","ahei3tert","nevfcur","alcohfreqwk",
"global_stress2", "apob_apoatert","whrs2tert","htnadmbp","cardiacrfcat","dmhba1c2")
in_out <- list(inlist=list(in_phys,in_ahei,in_nevfcur,in_alcohfreqwk,in_global_stress2,
in_htnadmbp, in_apob_apoatert,in_whrs2tert,in_cardiacrfcat,
in_dmhba1c2,in_case),
outlist=c("phys","ahei3tert","nevfcur","alcohfreqwk","global_stress2",
"htnadmbp","apob_apoatert", "whrs2tert","cardiacrfcat",
"dmhba1c2","case"))
if(checkMarkovDAG(in_out)$IsMarkovDAG & !checkMarkovDAG(in_out)$Reordered){
print("Your in_out DAG is a Markov DAG.")
} else if( checkMarkovDAG(in_out)$IsMarkovDAG & checkMarkovDAG(in_out)$Reordered ) {
in_out <- checkMarkovDAG(in_out)[[2]]
print("Your in_out DAG is a Markov DAG.The checkMarkovDAG function has reordered your
in_out list so that all parent variables come before descendants.")
} else{ print("Your ``in_out'' list is not a Bayesian Markov DAG so the methods in the
'causalPAF' package cannot be applied for non Markov DAGs.")}
w <- rep(1,nrow(stroke_reduced))
w[stroke_reduced$case==0] <- 0.9965
w[stroke_reduced$case==1] <- 0.0035
stroke_reduced$weights <- w
# 'NumOrderRiskFactors' should be set to a large number to ensure accurate results.
# This can take time to run.
sequentialPAF <- sequential_PAF( dataframe = stroke_reduced,
model_list_var = list(),
weights = w,
in_outDAG = in_out,
response = "case",
NumOrderRiskFactors = 3,
addCustom = TRUE,
custom = "regionnn7*ns(eage,df=5)+esex*ns(eage,df=5)" )
sequentialPAF$SAF_summary
#######################################################################################
# Alternatively, the user can supply a customised model_list_var parameter as follows:
# Libraries must be loaded if fitting models outside of the 'causalPAF' R package.
library(MASS)
library(splines)
# model_list_var is a list of models fitted for each of the variables in in_outDAG$outlist based
# on its parents given in in_outDAG$inlist. By default this is set to an empty list.
# Alternatively the user can supply their custom fitted, model_list as follows, which should be
# consistent with the causal structure.
# Note it is important that model_listArg is defined as a list and in the same order and length
# as the variables defined in in_outDAG[[2]].
model_list <- list(
glm(formula = phys ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) + moteduc
+ fatduc, family = "binomial", data = stroke_reduced, weights = weights), # model 1 phys
polr(formula = ahei3tert ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, data = stroke_reduced, weights = weights), # model 2 ahei3tert
glm(formula = nevfcur ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, family = "binomial",data = stroke_reduced, weights = weights), # model 3 nevfcur
polr(formula = alcohfreqwk ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, data = stroke_reduced,weights = weights), # model 4 alcohfreqwk
glm(formula = global_stress2 ~ subeduc + regionnn7 * ns(eage,df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc, family = "binomial",data = stroke_reduced,
weights = weights), # model 5 global_stress2
glm(formula = htnadmbp ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert + nevfcur + alcohfreqwk + global_stress2,
family = "binomial",data = stroke_reduced, weights = weights), # model 6 htnadmbp
polr(formula = apob_apoatert ~ regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
subeduc + moteduc + fatduc + phys + ahei3tert + nevfcur + alcohfreqwk + global_stress2,
data = stroke_reduced,weights = weights), # model 7 apob_apoatert
polr(formula = whrs2tert ~ regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) + subeduc +
moteduc + fatduc + phys + ahei3tert + nevfcur + alcohfreqwk + global_stress2,
data = stroke_reduced, weights = weights), # model 8 whrs2tert
glm(formula = cardiacrfcat ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert + nevfcur + alcohfreqwk + global_stress2 + apob_apoatert +
whrs2tert + htnadmbp, family = "binomial",
data = stroke_reduced, weights = weights), # model 9 cardiacrfcat
glm(formula = dmhba1c2 ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert + nevfcur + alcohfreqwk + global_stress2 + apob_apoatert +
whrs2tert + htnadmbp, family = "binomial",
data = stroke_reduced, weights = weights), # model 10 dmhba1c2
glm(formula = case ~ subeduc + regionnn7 * ns(eage, df = 5) + esex * ns(eage, df = 5) +
moteduc + fatduc + phys + ahei3tert + nevfcur + alcohfreqwk + global_stress2 + apob_apoatert +
whrs2tert + htnadmbp + cardiacrfcat + dmhba1c2, family = "binomial", data = stroke_reduced,
weights = weights) # model 11 case
)
# 'NumOrderRiskFactors' should be set to a large number to ensure accurate results.
# This can take time to run.
sequentialPAF <- sequential_PAF( dataframe = stroke_reduced,
model_list_var = model_list,
weights = stroke_reduced$weights,
in_outDAG = in_out,
response = "case",
NumOrderRiskFactors = 3 )
sequentialPAF$SAF_summary
Set a variable defined in col_num to its reference level (or otherwise 0 i.e. symbolising the absence of the risk factor) and then simulate all it's children.
Description
Set variable defined in col_num to its reference level or otherwise to 0. Then simulate each variable if there is a direct arrow from the variable thats set to its reference value (or otherwise 0) to that variable i.e. if the variable set to its reference level (or otherwise 0) is a parent of a variable simulate the child.
Usage
sim_outnode(dataframe, col_num, current_mat, in_outArg, col_list, model_list)
Arguments
dataframe |
A wide format dataframe containing all the risk factors, confounders, exposures and outcomes within the causal DAG Bayesian network. |
col_num |
Column number of variable in dataframe current_mat that is to be set to its reference level (if a factor) or to 0 otherwise i.e. the absence of the risk factor. |
current_mat |
Data frame containing the data that is to be manipulated by the function. The data frame has cases in rows and variables in columns. |
in_outArg |
This defines the causal directed acyclic graph (DAG). A list of length 2. It is defined as a two dimensional list consisting of, firstly, the first list, inlist, i.e. a list of the parents of each variable of interest corresponding to its column name in the data. Splines can be included here if they are to be modelled as splines. Secondly, the second list, outlist, contains a list of a single name of exposure or risk factor or outcome in form of characters i.e. a list of each variable of interest (risk factors, exposures and outcome) corresponding to its column name in the data. Splines should not be input here, only the column names of the variables of interest in the data. The order at which variables are defined must satisfy (i) It is important that variables are defined in the same order in both lists e.g. the first risk factor defined in outlist has its parents listed first in inlist, the second risk factor defined in outlist has its parents listed secondly in inlist and so on. The package assumes this ordering and will not work if this order is violated. (ii) Note it is important also that the order at which the variables are defined is such that all parents of that variable are defined before it. See example in tutorial. |
col_list |
is a list which gives the column numbers of each column in dataframe and current_mat that are in the same order as in_outArg and model_list. See tutorial for an example. |
model_list |
is a list of models fitted for each of the variables in in_outArg[[2]] (or in_outArg |
Value
current_mat |
Returns the dataframe current_mat updated by setting the exposure or variable in column, col_num, of current_mat to its reference level (if a factor) or to 0 otherwise i.e. the absence of the risk factor. current_mat is then returned with all children of column, col_num, of current_mat simulated given col_num, of current_mat has been set to its reference level (if a factor) or to 0 otherwise i.e. the absence of the risk factor. In summary, current_mat is returned setting the variable defined in col_num to its reference level (or otherwise 0 i.e. symbolising the absence of the risk factor) and then simulates all it's children based on this reference value of the variable defined at col_cum of current_mat. This allows for indirect effects and direct effects. |
Set a variable defined in col_num to its reference level (or otherwise 0 i.e. symbolising the absence of the risk factor) and then simulate all it's children.
Description
Set variable defined in col_num to its reference level or otherwise to 0. Then simulate each variable if there is a direct arrow from the variable thats set to its reference value (or otherwise 0) to that variable i.e. if the variable set to its reference level (or otherwise 0) is a parent of a variable simulate the child.
Usage
sim_outnode_2(
dataframe,
col_num,
current_mat,
col_list,
model_list,
response_col_num,
in_outArg
)
Arguments
dataframe |
A wide format dataframe containing all the risk factors, confounders, exposures and outcomes within the causal DAG Bayesian network. |
col_num |
Column number of variable in dataframe current_mat that is to be set to its reference level (if a factor) or to 0 otherwise i.e. the absence of the risk factor. |
current_mat |
Data frame containing the data. The data frame has cases in rows and variables in columns. |
col_list |
is a list which gives the column numbers of each column in dataframe and current_mat that are in the same order as in_outArg and model_list. See tutorial for an example. |
model_list |
is a list of models fitted for each of the variables in in_outArg[[2]] (or in_outArg |
response_col_num |
The column of current_mat and dataframe that defines the response variable. |
in_outArg |
This defines the causal directed acyclic graph (DAG). A list of length 2. It is defined as a two dimensional list consisting of, firstly, the first list, inlist, i.e. a list of the parents of each variable of interest corresponding to its column name in the data. Splines can be included here if they are to be modelled as splines. Secondly, the second list, outlist, contains a list of a single name of exposure or risk factor or outcome in form of characters i.e. a list of each variable of interest (risk factors, exposures and outcome) corresponding to its column name in the data. Splines should not be input here, only the column names of the variables of interest in the data. The order at which variables are defined must satisfy (i) It is important that variables are defined in the same order in both lists e.g. the first risk factor defined in outlist has its parents listed first in inlist, the second risk factor defined in outlist has its parents listed secondly in inlist and so on. The package assumes this ordering and will not work if this order is violated. (ii) Note it is important also that the order at which the variables are defined is such that all parents of that variable are defined before it. See example in tutorial. |
Value
current_mat |
Returns the dataframe current_mat updated by setting the exposure or variable in column, col_num, of current_mat to its reference level (if a factor) or to 0 otherwise i.e. the absence of the risk factor. current_mat is then returned with only the response model updated given col_num, of current_mat has been set to its reference level (if a factor) or to 0 otherwise i.e. the absence of the risk factor. In summary, current_mat is returned setting the variable defined in col_num to its reference level (or otherwise 0 i.e. symbolising the absence of the risk factor) and then simulates only the response model based on this reference value of the variable defined at col_cum of current_mat. This allows for only direct effects. There are no indirect effects simulated. |
Fictional ischemic stroke data case control data with risk factors, exposures and confounders
Description
A fictional standardized international study of ischemic stroke cases and controls in 32 countries in Asia, Europe, Australia, the Middle East and Africa. Information on key causal and modifiable risk factors for stroke are included. The risk factors included are healthy eating score (in tertiles), physical inactivity (yes/no), smoking behaviour (current smoker or ex/no smoker), alcohol intake (no alcohol, moderate consumption, high consumption), an indicator for stress, ApoB/ApoA lipid ratio (in tertiles), preexisting hypertension or high measured blood pressure (yes/no), waist hip ratio (in tertiles), cardiac risk factors such as atrial fibrillation or flutter (yes or no) and a diagnosis of diabetes mellitus or elevated HbA1c(yes/no). One needs to assume a causal graph describing the causal relationships between confounders, risk factors and disease to implement the methods in the 'causalPAF' package. To do this, it is helpful to divide the risk factors and exposures into categories depending on whether they are descriptive of an individual’s behaviour SB = Smoking, Alcohol intake, inactivity, diet and stress, their physiology SP = High blood pressure, ApoB/ApoA ratio, Waist hip ratio and what might be regarded as preclinical disease SD = Cardiac risk factors, Preclinical diabetes. We also consider a set of variables that might be confounders (joint causes of the risk factor and stroke) for all the listed risk factors. This set of confounders, SC consist of the individuals and their parents’ education level (in 5 levels from no education to holding a college Degree), age, gender and region. Here we make the simplifying assumption that disease develops in a stage wise fashion, each stage being represented by one of the sets of variables just described with variables in earlier stages having causal effects on variables contained in later stages, but not vice versa. The ordering of stages is indicated by the sequence, SC , SB , SP , SD , Y , and summarized by the causal graphs in (O’Connell and Ferguson 2020) <https://doi.org/10.1101/2020.10.15.20212845> and (Ferguson, O’Connell, and O’Donnell 2020) Revisiting sequential attributable fractions, J Ferguson, M O'Connell, M O'Donnell, Archives of Public Health, 2020.
Usage
strokedata
Format
A data frame with 16623 rows and 21 variables in columns. Each row representing either a fictional individual stroke case or control:
- regionnn7
Region (number, 7 categories)
- case
Case status (number, 1=cases, 0=controls)
- esex
Gender (number, 1=female, 2=male)
- eage
,Age based on both DOB and eage (number)
- htnadmbp
High blood pressure, Hx HTN/Adjusted BP>140/90 at admission (number, 0=No, 1=Yes)
- nevfcur
Smoke history(2lev) (number, 1=Never/Former, 2=Current)
- global_stress2
Global Stress, (number, 1=None/some_periods, 2=Several/Perm. periods)
- whrs2tert
Tertile of standing waist to hip ratio, WHR (number, 1=Tertile1, 2=Tertile2, 3=Tertile3)
- phys
Leisure Physical activity (number, 1=mainly inactive, 2=mainly active)
- alcohfreqwk
Alcohol history and frequency(3) (number, 1=Never/Former, 2=Low/Moderate, 3=High intake/Binge)
- dmhba1c2
clinically diagnosed diabetes mellitus or measured Hba1c level at least 6.5 yes or no, Hx DM/HbA1c greater than or equal to 6.5 per cent (number, 1=No, 2=Yes)
- cardiacrfcat
History of risk factors for heart disease yes or no, Hx Cardiac Risk Factors (number, 1=No, 2=Yes)
- ahei3tert
Diet: AHEI diet score (in tertiles) Tertile of Total AHEI Score (not including alcohol), (number, 1=Tertile1, 2=Tertile2, 3=Tertile3)
- apob_apoatert
Lipids: Apolipoprotein B/Apolipoprotein A1 ratio (in tertiles) (number, 1=Tertile1, 2=Tertile2, 3=Tertile3)
- subeduc
Education (number, 1=None, 2=1 to 8, 3=9 to 12, 4=Trade School, 5=College/University)
- moteduc
Mother education (number, 1=None, 2=1 to 8, 3=9 to 12, 4=Trade School, 5=College/University, 6=unknown)
- fatduc
Father education (number, 1=None, 2=1 to 8, 3=9 to 12, 4=Trade School, 5=College/University, 6=Unknown)
- subhtn
Blood pressure Hx HTN (number, 1=No, 2=Yes)
- whr
Waist to hip ratio ( number as continuous variable)
- apob_apoa
Lipids: Apolipoprotein B/Apolipoprotein A1 ratio (number as continuous variable)
- weights
To fit these models to case control data, one needs to perform weighted maximum likelihood estimation to imitate estimation using a random sample from the population. We chose weights of 0.0035 (for each case) and 0.9965 (for each control), reflective of a yearly incidence of first ischemic stroke of 0.35 per cent, or 3.5 strokes per 1,000 individuals. These weights were chosen according to average incidences across country, age group and gender according to the global burden of disease.
Fictional ischemic stroke data case control data with risk factors, exposures and confounders
Description
A fictional standardized international study of ischemic stroke cases and controls in 32 countries in Asia, Europe, Australia, the Middle East and Africa. Information on key causal and modifiable risk factors for stroke are included. The risk factors included are healthy eating score (in tertiles), physical inactivity (yes/no), smoking behaviour (current smoker or ex/no smoker), alcohol intake (no alcohol, moderate consumption, high consumption), an indicator for stress, ApoB/ApoA lipid ratio (in tertiles), pre existing hypertension or high measured blood pressure (yes/no), waist hip ratio (in tertiles), cardiac risk factors such as atrial fibrillation or flutter (yes or no) and a diagnosis of diabetes mellitus or elevated HbA1c(yes/no). One needs to assume a causal graph describing the causal relationships between confounders, risk factors and disease to implement the methods in the 'causalPAF' package. To do this, it is helpful to divide the risk factors and exposures into categories depending on whether they are descriptive of an individual’s behaviour SB = Smoking, Alcohol intake, inactivity, diet and stress, their physiology SP = High blood pressure, ApoB/ApoA ratio, Waist hip ratio and what might be regarded as preclinical disease SD = Cardiac risk factors, Pre clinical diabetes. We also consider a set of variables that might be confounders (joint causes of the risk factor and stroke) for all the listed risk factors. This set of confounders, SC consist of the individuals and their parents’ education level (in 5 levels from no education to holding a college Degree), age, gender and region. Here we make the simplifying assumption that disease develops in a stage wise fashion, each stage being represented by one of the sets of variables just described with variables in earlier stages having causal effects on variables contained in later stages, but not vice versa. The ordering of stages is indicated by the sequence, SC , SB , SP , SD , Y , and summarized by the causal graphs in (O’Connell and Ferguson 2020) <https://doi.org/10.1101/2020.10.15.20212845> and (Ferguson, O’Connell, and O’Donnell 2020) Revisiting sequential attributable fractions, J Ferguson, M O'Connell, M O'Donnell, Archives of Public Health, 2020..
Usage
strokedata
Format
A data frame with 5000 rows and 21 variables in columns. Each row representing either a fictional individual stroke case or control:
- regionnn7
Region (number, 7 categories)
- case
Case status (number, 1=cases, 0=controls)
- esex
Gender (number, 1=female, 2=male)
- eage
,Age based on both DOB and eage (number)
- htnadmbp
High blood pressure, Hx HTN/Adjusted BP>140/90 at admission (number, 0=No, 1=Yes)
- nevfcur
Smoke history(2lev) (number, 1=Never/Former, 2=Current)
- global_stress2
Global Stress, (number, 1=None/some_periods, 2=Several/Perm. periods)
- whrs2tert
Tertile of standing waist to hip ratio, WHR (number, 1=Tertile1, 2=Tertile2, 3=Tertile3)
- phys
Leisure Physical activity (number, 1=mainly inactive, 2=mainly active)
- alcohfreqwk
Alcohol history and frequency(3) (number, 1=Never/Former, 2=Low/Moderate, 3=High intake/Binge)
- dmhba1c2
clinically diagnosed diabetes mellitus or measured Hba1c level at least 6.5 yes or no, Hx DM/HbA1c greater than or equal to 6.5 per cent (number, 1=No, 2=Yes)
- cardiacrfcat
History of risk factors for heart disease yes or no, Hx Cardiac Risk Factors (number, 1=No, 2=Yes)
- ahei3tert
Diet: AHEI diet score (in tertiles) Tertile of Total AHEI Score (not including alcohol), (number, 1=Tertile1, 2=Tertile2, 3=Tertile3)
- apob_apoatert
Lipids: Apolipoprotein B/Apolipoprotein A1 ratio (in tertiles) (number, 1=Tertile1, 2=Tertile2, 3=Tertile3)
- subeduc
Education (number, 1=None, 2=1 to 8, 3=9 to 12, 4=Trade School, 5=College/University)
- moteduc
Mother education (number, 1=None, 2=1 to 8, 3=9 to 12, 4=Trade School, 5=College/University, 6=unknown)
- fatduc
Father education (number, 1=None, 2=1 to 8, 3=9 to 12, 4=Trade School, 5=College/University, 6=Unknown)
- subhtn
Blood pressure Hx HTN (number, 1=No, 2=Yes)
- whr
Waist to hip ratio ( number as continuous variable)
- apob_apoa
Lipids: Apolipoprotein B/Apolipoprotein A1 ratio (number as continuous variable)
- weights
To fit these models to case control data, one needs to perform weighted maximum likelihood estimation to imitate estimation using a random sample from the population. We chose weights of 0.0035 (for each case) and 0.9965 (for each control), reflective of a yearly incidence of first ischemic stroke of 0.35 per cent, or 3.5 strokes per 1,000 individuals. These weights were chosen according to average incidences across country, age group and gender according to the global burden of disease.